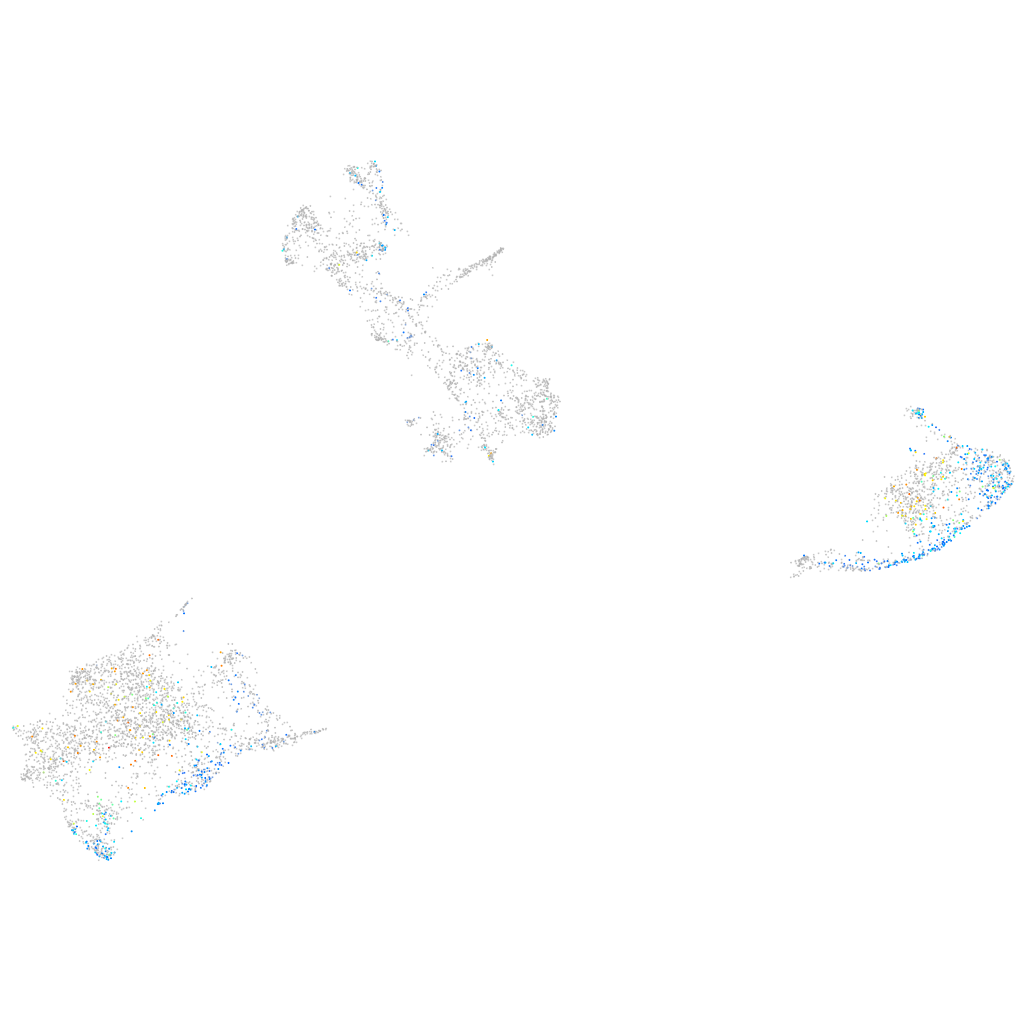
si:dkey-24c2.9
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia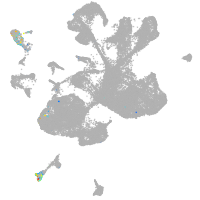 |
eye |
taste / olfactory |
otic / lateral line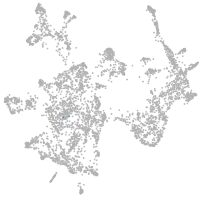 |
epidermis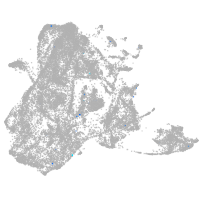 |
periderm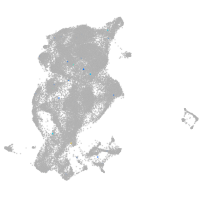 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tspan36 | 0.202 | defbl1 | -0.103 |
| si:ch73-389b16.1 | 0.201 | mdkb | -0.097 |
| slc24a5 | 0.195 | si:ch73-1a9.3 | -0.097 |
| dct | 0.193 | apoda.1 | -0.096 |
| tyrp1a | 0.193 | hmgn2 | -0.092 |
| tyrp1b | 0.189 | si:ch211-222l21.1 | -0.092 |
| pmela | 0.185 | gpnmb | -0.089 |
| tspan10 | 0.185 | si:ch211-256m1.8 | -0.089 |
| tyr | 0.182 | lypc | -0.089 |
| zgc:91968 | 0.180 | si:dkey-197i20.6 | -0.082 |
| mlpha | 0.177 | tuba1c | -0.082 |
| oca2 | 0.172 | alx4a | -0.079 |
| slc3a2a | 0.170 | ptmab | -0.077 |
| tfap2e | 0.169 | ptmaa | -0.076 |
| mitfa | 0.166 | s100v2 | -0.075 |
| mtbl | 0.162 | h2afvb | -0.074 |
| slc7a5 | 0.159 | hnrnpaba | -0.074 |
| gstt1a | 0.158 | slc25a36a | -0.073 |
| slc45a2 | 0.158 | akap12a | -0.073 |
| pttg1ipb | 0.156 | gpm6aa | -0.073 |
| tmem243b | 0.153 | si:ch73-89b15.3 | -0.073 |
| slc22a2 | 0.151 | stmn1b | -0.072 |
| rasd1 | 0.151 | alx1 | -0.072 |
| slc24a4a | 0.150 | hmga1a | -0.071 |
| inka1b | 0.150 | pltp | -0.071 |
| kita | 0.150 | hmgb1b | -0.070 |
| pcbd1 | 0.149 | unm-sa821 | -0.070 |
| si:ch211-195b13.1 | 0.146 | pcna | -0.069 |
| csrp2 | 0.146 | aqp1a.1 | -0.069 |
| rab38 | 0.145 | tuba1b | -0.069 |
| aadac | 0.144 | prx | -0.069 |
| rab34b | 0.144 | stmn1a | -0.068 |
| slc37a2 | 0.144 | ponzr1 | -0.068 |
| SPAG9 | 0.143 | elavl3 | -0.068 |
| kcnj13 | 0.143 | cx43 | -0.068 |