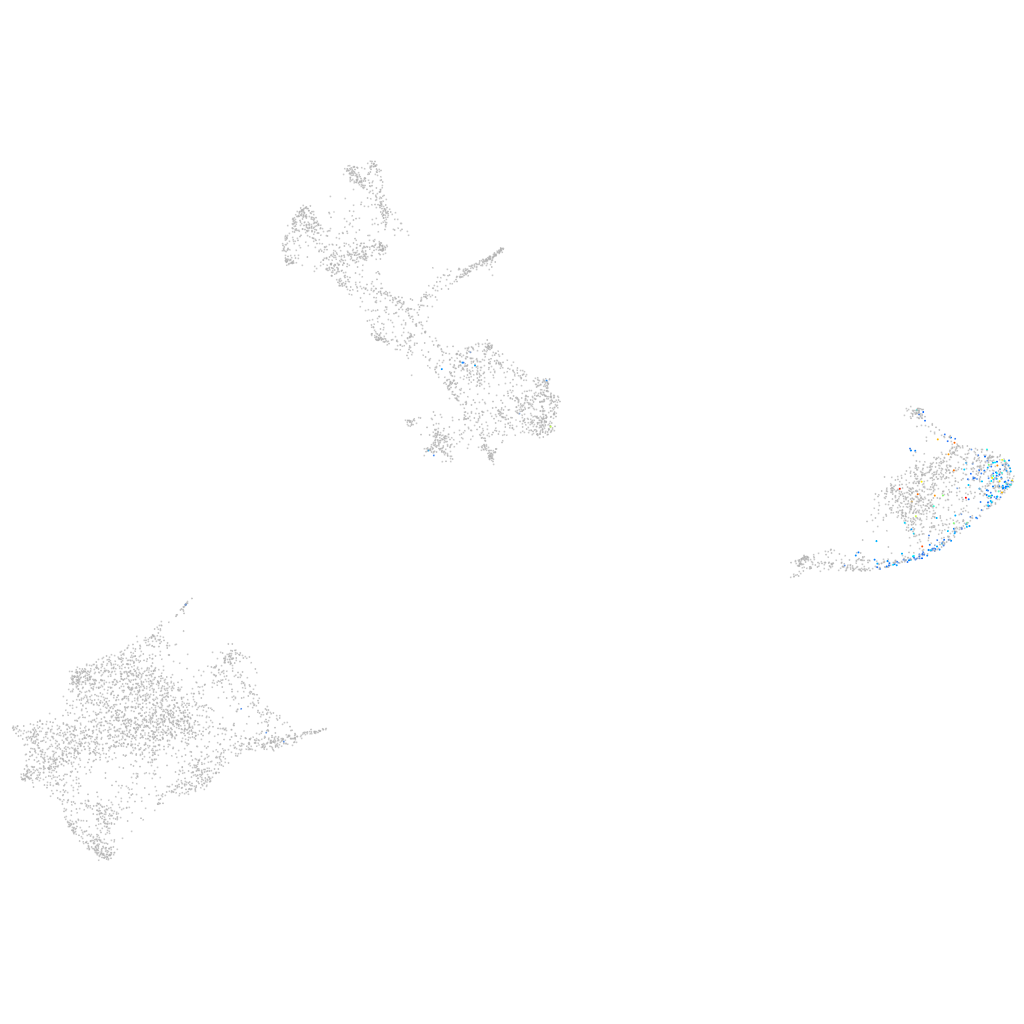
si:dkey-247m21.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kita | 0.312 | paics | -0.150 |
| tyrp1a | 0.305 | CABZ01021592.1 | -0.149 |
| oca2 | 0.304 | uraha | -0.141 |
| tyrp1b | 0.300 | si:dkey-251i10.2 | -0.122 |
| pmela | 0.296 | mdh1aa | -0.116 |
| aadac | 0.290 | tmem130 | -0.113 |
| slc24a5 | 0.289 | slc2a15a | -0.109 |
| dct | 0.286 | atic | -0.105 |
| prkar1b | 0.275 | glulb | -0.103 |
| zgc:91968 | 0.275 | impdh1b | -0.100 |
| slc39a10 | 0.274 | aox5 | -0.097 |
| SPAG9 | 0.274 | phyhd1 | -0.097 |
| si:ch73-389b16.1 | 0.272 | si:ch211-251b21.1 | -0.095 |
| slc7a5 | 0.258 | prps1a | -0.088 |
| mchr2 | 0.253 | bscl2l | -0.087 |
| tyr | 0.252 | krt18b | -0.084 |
| lamp1a | 0.250 | pax7b | -0.082 |
| mtbl | 0.248 | pax7a | -0.079 |
| slc22a2 | 0.242 | bco1 | -0.077 |
| slc45a2 | 0.241 | cyb5a | -0.077 |
| slc29a3 | 0.238 | ppat | -0.077 |
| kcnj13 | 0.235 | shmt1 | -0.074 |
| hsd20b2 | 0.232 | sult1st1 | -0.073 |
| gpr61 | 0.229 | cx30.3 | -0.073 |
| slc30a1b | 0.228 | sprb | -0.073 |
| slc30a8 | 0.226 | cax1 | -0.072 |
| anxa1a | 0.222 | defbl1 | -0.072 |
| tfap2e | 0.219 | mibp | -0.072 |
| CDK18 | 0.217 | si:ch211-137a8.4 | -0.071 |
| zdhhc2 | 0.215 | apoda.1 | -0.071 |
| gstt1a | 0.214 | pmp22b | -0.069 |
| mlpha | 0.213 | aldob | -0.069 |
| slc6a15 | 0.213 | oacyl | -0.068 |
| rap1gap | 0.209 | hmgn2 | -0.068 |
| slc37a2 | 0.209 | rbp4l | -0.067 |