si:dkey-245n4.2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 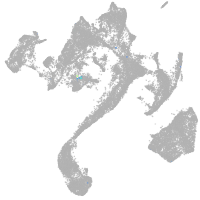 |
mural cells / non-skeletal muscle 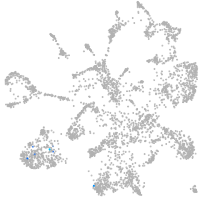 |
mesenchyme 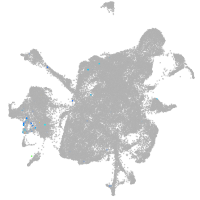 |
hematopoietic / vasculature 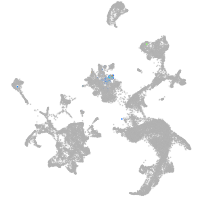 |
pronephros  |
neural |
spinal cord / glia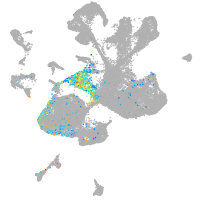 |
eye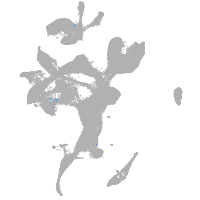 |
taste / olfactory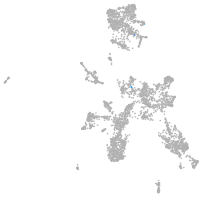 |
otic / lateral line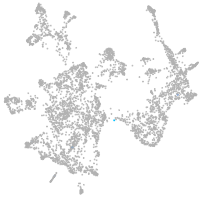 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cldn7a | 0.488 | hbbe3 | -0.021 |
| slc6a11b | 0.427 | cdh5 | -0.018 |
| gfap | 0.419 | si:ch211-156j16.1 | -0.018 |
| hepacama | 0.393 | etv2 | -0.018 |
| entpd2b | 0.384 | CT030188.1 | -0.018 |
| atp1b4 | 0.342 | she | -0.017 |
| zgc:165461 | 0.330 | yrk | -0.017 |
| slc10a1 | 0.328 | srgn | -0.017 |
| pkd1b | 0.328 | f13a1b | -0.017 |
| slc4a4a | 0.326 | plk2b | -0.017 |
| atp1a1b | 0.324 | dusp5 | -0.017 |
| si:ch73-265d7.2 | 0.308 | hhex | -0.017 |
| CR931782.2 | 0.296 | clec14a | -0.016 |
| slc1a2b | 0.281 | tal1 | -0.016 |
| hspb15 | 0.280 | rac2 | -0.016 |
| clstn2 | 0.279 | myct1a | -0.016 |
| her8.2 | 0.277 | rab5c | -0.016 |
| heyl | 0.276 | b3gnt3.4 | -0.016 |
| endouc | 0.267 | blf | -0.016 |
| slc7a10a | 0.267 | hmbsb | -0.016 |
| cfap161 | 0.257 | plvapb | -0.016 |
| gpr37l1b | 0.247 | tmem88a | -0.016 |
| rfx4 | 0.242 | tagln2 | -0.016 |
| kcnn1b | 0.241 | tie1 | -0.016 |
| cfap157 | 0.239 | mcamb | -0.015 |
| zgc:153704 | 0.233 | XLOC-014079 | -0.015 |
| si:ch211-251b21.1 | 0.232 | ehd4 | -0.015 |
| si:ch211-66e2.5 | 0.229 | ecscr | -0.015 |
| si:ch1073-303k11.2 | 0.228 | pecam1 | -0.015 |
| lrrc39 | 0.226 | pfn1 | -0.015 |
| vwa7 | 0.225 | csrp1a | -0.015 |
| slc1a3b | 0.224 | si:ch73-248e21.7 | -0.015 |
| XLOC-024343 | 0.223 | si:dkey-52l18.4 | -0.015 |
| eno1b | 0.220 | jpt2 | -0.015 |
| ptgdsb.2 | 0.217 | si:dkeyp-97a10.2 | -0.015 |