si:dkey-204f11.64
ZFIN 
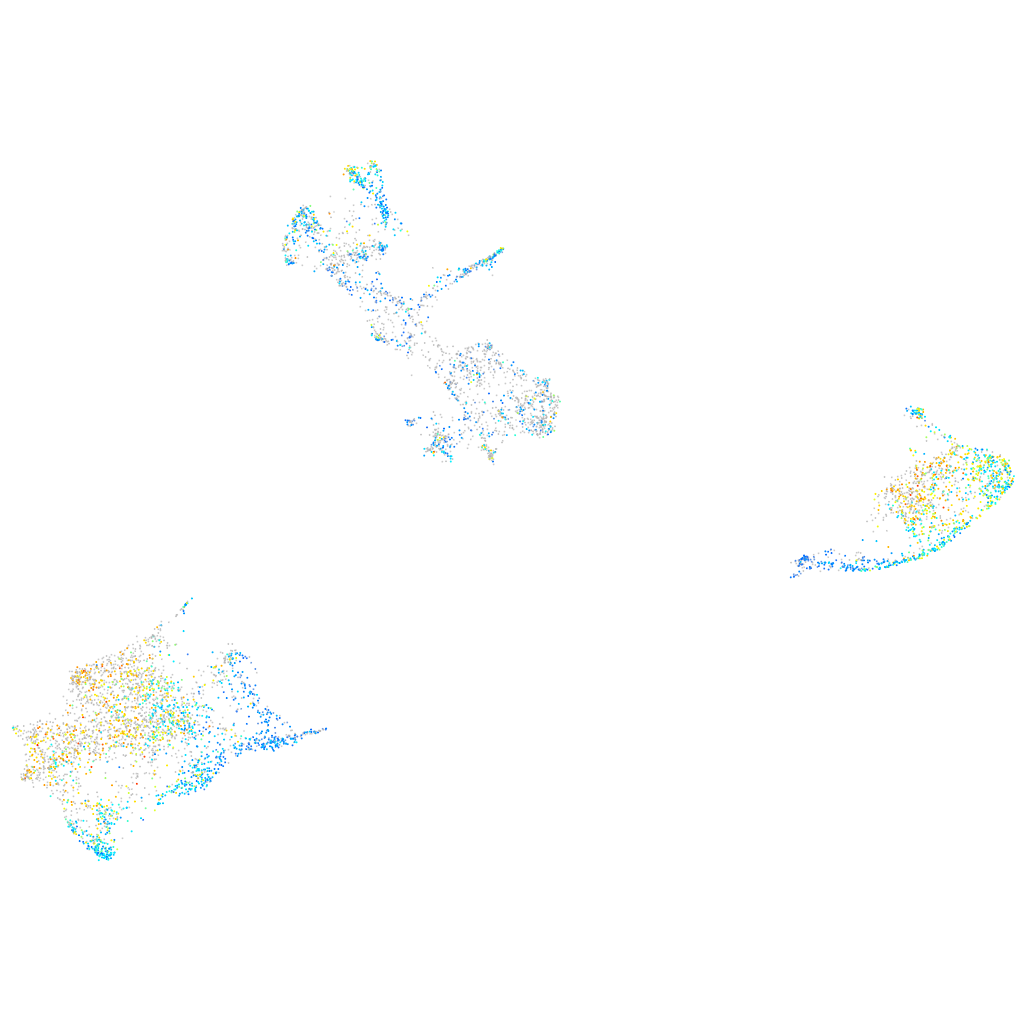
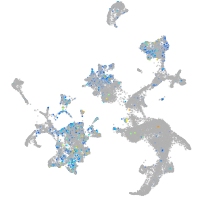
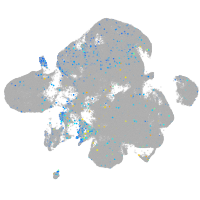
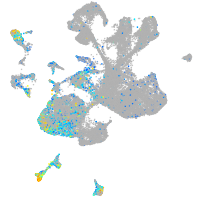
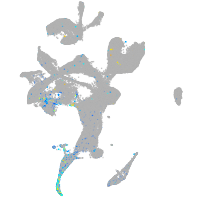
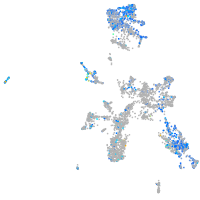
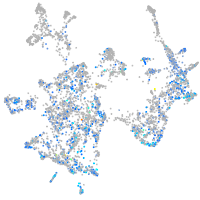
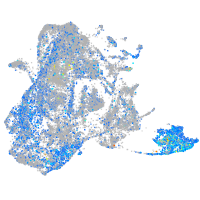
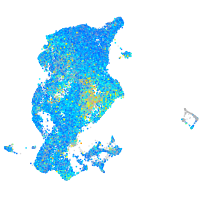
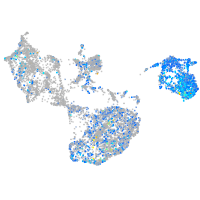
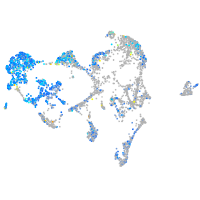
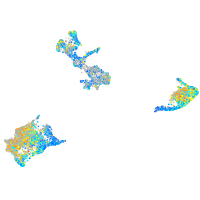
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-389b16.1 | 0.260 | si:ch211-222l21.1 | -0.240 |
| tspan36 | 0.259 | mdkb | -0.181 |
| mlpha | 0.248 | hmgn2 | -0.178 |
| slc24a5 | 0.246 | si:ch73-1a9.3 | -0.174 |
| tspan10 | 0.243 | ptmaa | -0.174 |
| zgc:91968 | 0.243 | ptmab | -0.165 |
| slc45a2 | 0.241 | gpm6aa | -0.164 |
| pmela | 0.241 | hmgb2b | -0.162 |
| tyrp1b | 0.239 | nova2 | -0.161 |
| tyrp1a | 0.239 | hmgb1b | -0.156 |
| slc3a2a | 0.233 | tmsb | -0.150 |
| oca2 | 0.230 | si:ch73-281n10.2 | -0.148 |
| dct | 0.228 | elavl3 | -0.147 |
| rab38 | 0.227 | hnrnpaba | -0.147 |
| s100a10b | 0.225 | stmn1a | -0.145 |
| prkar1b | 0.219 | setb | -0.144 |
| kita | 0.219 | tuba1c | -0.143 |
| pcbd1 | 0.219 | tmeff1b | -0.141 |
| kcnj13 | 0.217 | hmgb3a | -0.140 |
| ucp2 | 0.217 | gng3 | -0.138 |
| gstt1a | 0.215 | si:ch211-288g17.3 | -0.138 |
| tfap2e | 0.213 | h2afvb | -0.136 |
| slc22a2 | 0.213 | h3f3a | -0.134 |
| tyr | 0.213 | hmga1a | -0.134 |
| vamp3 | 0.212 | hmgb2a | -0.133 |
| pah | 0.212 | stmn1b | -0.133 |
| bace2 | 0.212 | epb41a | -0.132 |
| pttg1ipb | 0.211 | seta | -0.131 |
| rabl6b | 0.210 | apex1 | -0.130 |
| mtbl | 0.210 | pcna | -0.129 |
| anxa1a | 0.208 | CU467822.1 | -0.128 |
| cd63 | 0.207 | sox11a | -0.125 |
| csrp2 | 0.207 | dut | -0.123 |
| actb2 | 0.203 | hnrnpabb | -0.123 |
| gramd2aa | 0.200 | marcksl1a | -0.122 |