si:dkey-19b23.14
ZFIN 
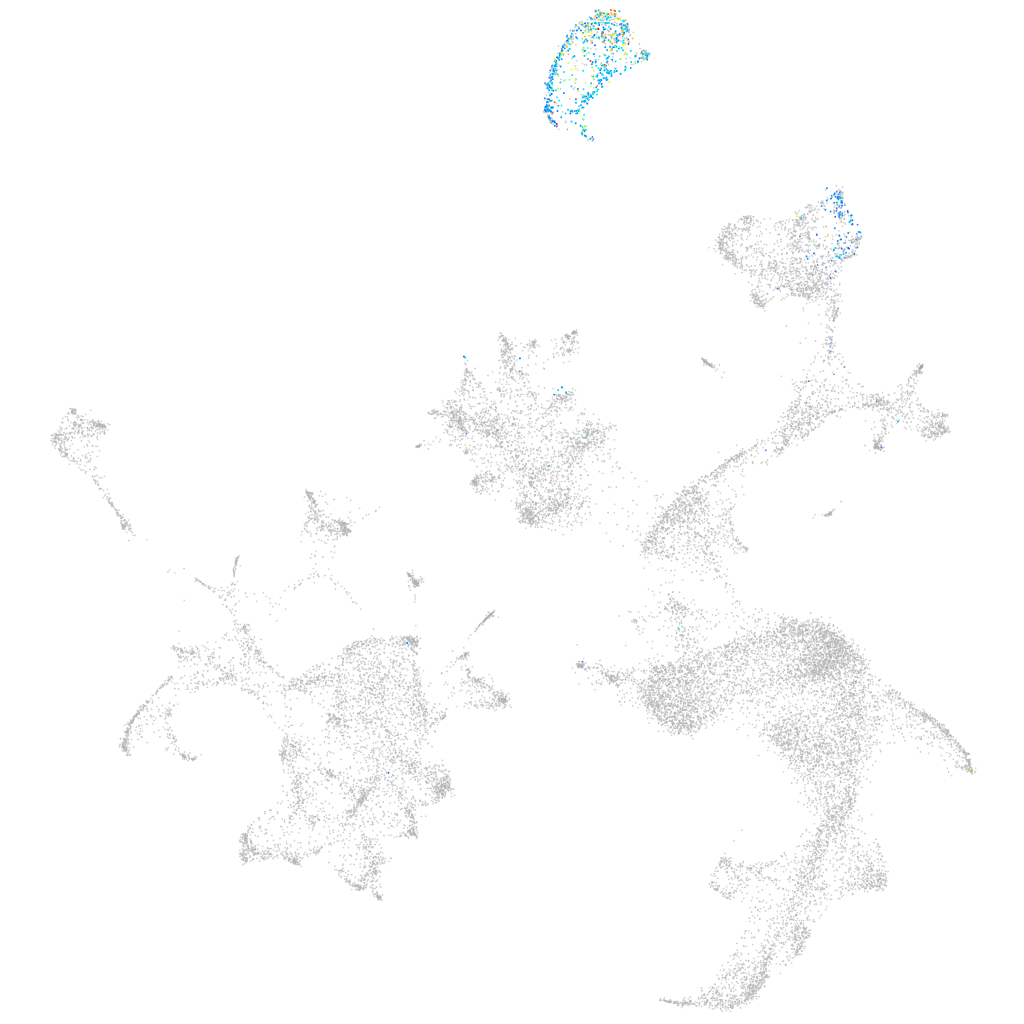
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npsn | 0.668 | hbae1.1 | -0.166 |
| lyz | 0.657 | hbae3 | -0.160 |
| mpx | 0.653 | hbbe1.3 | -0.160 |
| ch25hl2 | 0.643 | hbae1.3 | -0.157 |
| mmp13a | 0.641 | hbbe2 | -0.156 |
| il34 | 0.598 | hbbe1.1 | -0.154 |
| cpa5 | 0.596 | cahz | -0.142 |
| ponzr6 | 0.593 | lmo2 | -0.135 |
| si:ch1073-429i10.1 | 0.592 | hbbe1.2 | -0.135 |
| lect2l | 0.587 | hemgn | -0.133 |
| si:dkey-102g19.3 | 0.582 | blvrb | -0.133 |
| cfbl | 0.577 | prdx2 | -0.132 |
| scpp8 | 0.568 | alas2 | -0.125 |
| alox5ap | 0.556 | si:ch211-250g4.3 | -0.125 |
| scinlb | 0.554 | fth1a | -0.124 |
| abcb9 | 0.546 | creg1 | -0.123 |
| il6r | 0.544 | zgc:56095 | -0.122 |
| ctss1 | 0.543 | slc4a1a | -0.118 |
| lta4h | 0.535 | nt5c2l1 | -0.115 |
| prkcda | 0.535 | epb41b | -0.111 |
| sms | 0.532 | zgc:163057 | -0.111 |
| glipr1a | 0.523 | si:ch211-207c6.2 | -0.105 |
| si:ch211-284o19.8 | 0.519 | nmt1b | -0.102 |
| dhrs9 | 0.517 | aqp1a.1 | -0.101 |
| LOC101884267 | 0.513 | ctsla | -0.101 |
| si:dkey-27h10.2 | 0.511 | plac8l1 | -0.097 |
| ppdpfa | 0.509 | tmod4 | -0.096 |
| si:ch211-276a23.5 | 0.507 | si:ch211-227m13.1 | -0.091 |
| gapdh | 0.506 | hbae5 | -0.090 |
| nccrp1 | 0.505 | sptb | -0.089 |
| CR354540.1 | 0.503 | uros | -0.089 |
| aldh8a1 | 0.501 | selenoj | -0.088 |
| si:ch211-286b5.8 | 0.500 | tfr1a | -0.088 |
| vamp8 | 0.493 | znfl2a | -0.085 |
| si:ch73-359m17.5 | 0.479 | rfesd | -0.085 |