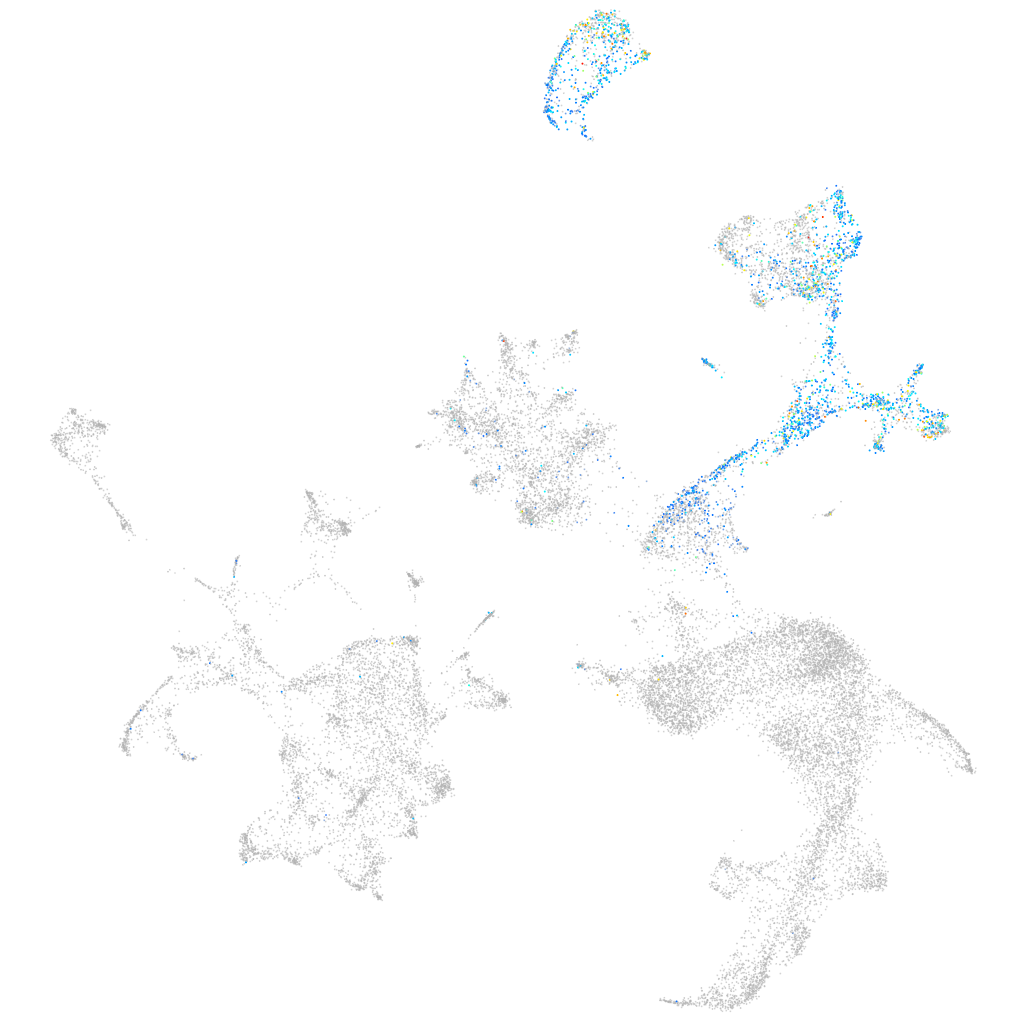
si:dkey-185m8.2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| coro1a | 0.490 | hbae3 | -0.268 |
| rac2 | 0.457 | hbbe1.3 | -0.264 |
| laptm5 | 0.456 | hbae1.3 | -0.258 |
| cebpb | 0.452 | hbbe2 | -0.254 |
| pfn1 | 0.452 | hbbe1.1 | -0.251 |
| grap2b | 0.435 | hbae1.1 | -0.246 |
| arpc1b | 0.432 | cahz | -0.236 |
| arhgdig | 0.431 | hbbe1.2 | -0.224 |
| wasb | 0.425 | alas2 | -0.211 |
| glipr1a | 0.410 | hemgn | -0.210 |
| wasa | 0.409 | si:ch211-250g4.3 | -0.201 |
| dub | 0.405 | creg1 | -0.200 |
| si:dkey-262k9.4 | 0.400 | blvrb | -0.195 |
| itgae.2 | 0.385 | nt5c2l1 | -0.193 |
| fmnl1a | 0.382 | fth1a | -0.192 |
| grap2a | 0.381 | zgc:56095 | -0.185 |
| ptpn6 | 0.378 | zgc:163057 | -0.184 |
| fam49ba | 0.377 | slc4a1a | -0.176 |
| ccr9a | 0.373 | si:ch211-207c6.2 | -0.174 |
| cotl1 | 0.372 | aqp1a.1 | -0.172 |
| spi1b | 0.371 | epb41b | -0.169 |
| eno3 | 0.367 | tspo | -0.161 |
| si:ch211-214p16.1 | 0.365 | tmod4 | -0.161 |
| ptprc | 0.364 | lmo2 | -0.155 |
| si:dkey-102g19.3 | 0.361 | nmt1b | -0.150 |
| zgc:64051 | 0.360 | hbae5 | -0.150 |
| hmha1b | 0.357 | si:ch211-227m13.1 | -0.147 |
| parvg | 0.355 | hbbe3 | -0.147 |
| pgd | 0.355 | plac8l1 | -0.144 |
| lpar5a | 0.354 | krt18a.1 | -0.143 |
| FERMT3 (1 of many) | 0.354 | sptb | -0.133 |
| nrros | 0.353 | si:dkey-240n22.3 | -0.132 |
| fcer1gl | 0.353 | uros | -0.131 |
| cd7al | 0.352 | akap12b | -0.128 |
| itgb2 | 0.350 | rfesd | -0.126 |