si:dkey-165o8.1
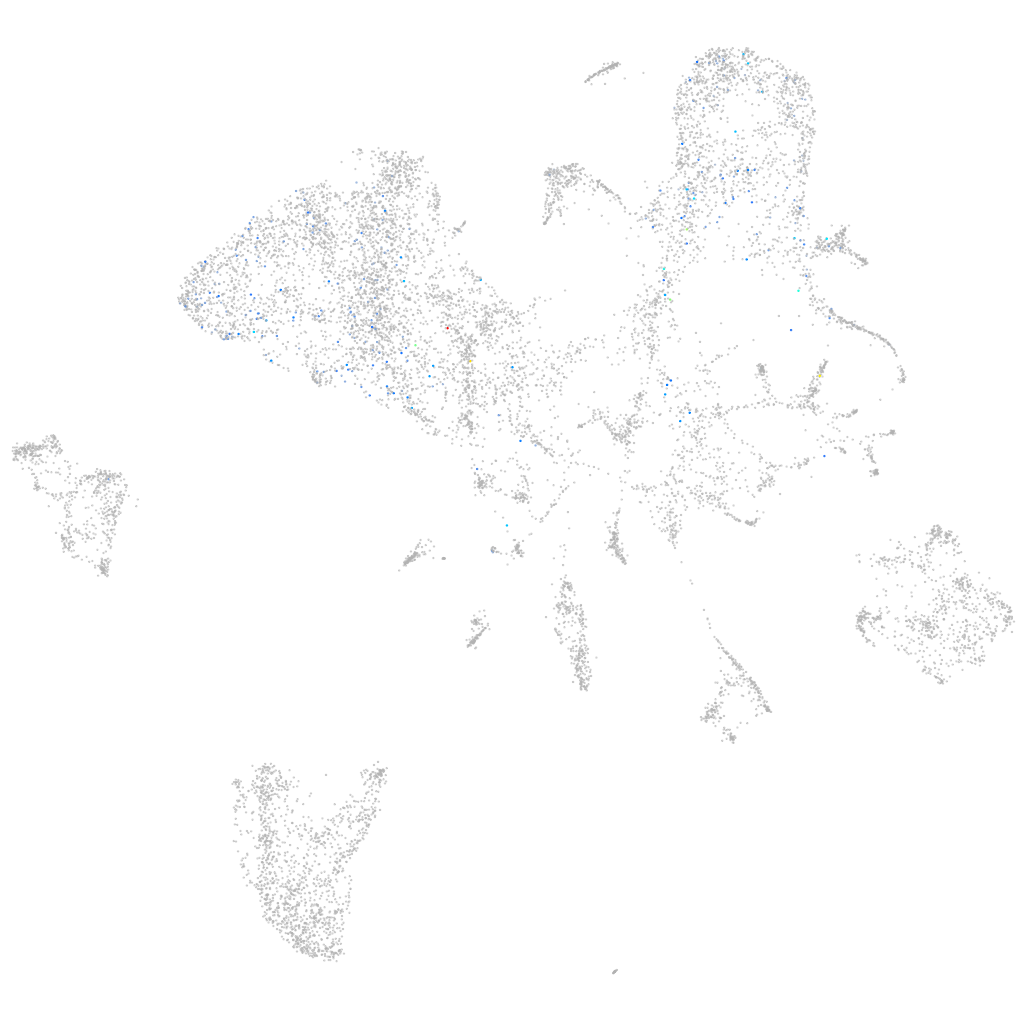
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory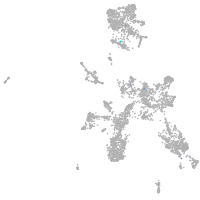 |
otic / lateral line |
epidermis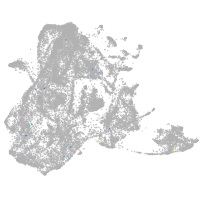 |
periderm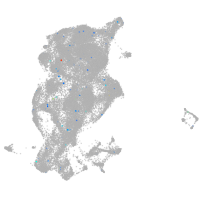 |
fin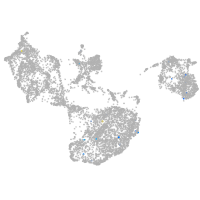 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC103911631 | 0.188 | si:ch211-195b11.3 | -0.055 |
| LOC110437921 | 0.153 | rtn1a | -0.052 |
| zgc:158427 | 0.148 | eef1a1l1 | -0.051 |
| LOC101883820 | 0.131 | slc38a5b | -0.050 |
| shisa6 | 0.119 | ctrl | -0.048 |
| taar20f | 0.117 | CELA1 (1 of many) | -0.047 |
| kif21b | 0.116 | ela3l | -0.045 |
| FP016005.1 | 0.115 | cpa5 | -0.045 |
| RF00007 | 0.111 | ela2 | -0.045 |
| si:ch211-207c6.2 | 0.110 | prss59.1 | -0.045 |
| CSRNP3 | 0.109 | prss1 | -0.045 |
| CU466287.2 | 0.107 | fep15 | -0.045 |
| kcnab2a | 0.104 | ela2l | -0.045 |
| sstr1a | 0.103 | ctrb1 | -0.045 |
| CABZ01088864.1 | 0.100 | erp27 | -0.045 |
| ompb | 0.099 | zgc:112160 | -0.045 |
| taar20i | 0.098 | prss59.2 | -0.044 |
| sult2st2 | 0.097 | sycn.2 | -0.044 |
| map3k7cl | 0.097 | pdia2 | -0.044 |
| gjd1a | 0.096 | cel.1 | -0.044 |
| hmx4 | 0.095 | cpb1 | -0.044 |
| LOC101882378 | 0.092 | si:ch211-240l19.5 | -0.043 |
| slco1d1 | 0.092 | cel.2 | -0.043 |
| aco1 | 0.092 | c6ast4 | -0.043 |
| eno3 | 0.092 | cpa4 | -0.043 |
| bgna | 0.092 | apoda.2 | -0.042 |
| sstr1b | 0.091 | amy2a | -0.042 |
| adtrp1 | 0.091 | zgc:136461 | -0.042 |
| hadh | 0.091 | npdc1a | -0.042 |
| hspd1 | 0.091 | cfl1l | -0.041 |
| LOC103911090 | 0.091 | marcksb | -0.041 |
| ndufs4 | 0.091 | endou | -0.041 |
| atp5mc1 | 0.090 | stard10 | -0.041 |
| suclg1 | 0.090 | zgc:92137 | -0.041 |
| creb3l3a | 0.090 | marcksl1a | -0.040 |