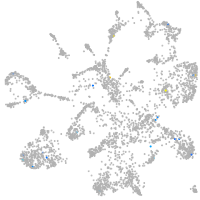
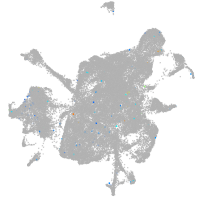
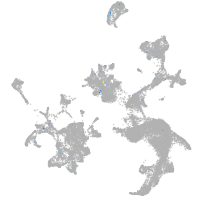
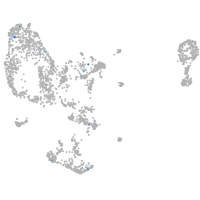
si:dkey-15h8.17
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| bmp10 | 0.206 | zgc:92429 | -0.021 |
| LO018644.1 | 0.175 | tpma | -0.020 |
| CU394253.1 | 0.168 | ggctb | -0.019 |
| ponzr10 | 0.167 | tcea3 | -0.018 |
| trim35-34 | 0.163 | tnnc2 | -0.018 |
| si:dkey-243k1.3 | 0.138 | dars | -0.018 |
| BX276103.1 | 0.132 | rps12 | -0.017 |
| zgc:174917 | 0.113 | rpl7a | -0.017 |
| lrrc34 | 0.104 | nop10 | -0.017 |
| slc24a4b | 0.104 | si:ch211-217k17.7 | -0.017 |
| CABZ01041494.1 | 0.101 | rwdd1 | -0.017 |
| p2ry10 | 0.101 | ddx27 | -0.016 |
| LOC101883088 | 0.100 | rpl6 | -0.016 |
| cacnb4a | 0.097 | hspb1 | -0.016 |
| terb1 | 0.090 | mrpl20 | -0.016 |
| ifnphi2 | 0.082 | hint1 | -0.016 |
| cfap57 | 0.082 | aamp | -0.016 |
| LOC108179562 | 0.082 | COX5B | -0.016 |
| inpp5jb | 0.080 | tsr1 | -0.016 |
| fam102aa | 0.078 | npm1a | -0.016 |
| myo3a | 0.076 | eif5a | -0.016 |
| XLOC-007906 | 0.074 | myoz1b | -0.015 |
| zgc:56231 | 0.073 | myl1 | -0.015 |
| ryr2b | 0.072 | bcas2 | -0.015 |
| si:dkey-247m21.3 | 0.071 | foxd3 | -0.015 |
| kiaa1549lb | 0.070 | trim55a | -0.015 |
| si:dkeyp-26a9.7 | 0.069 | hjv | -0.015 |
| XLOC-029420 | 0.069 | edf1 | -0.015 |
| marco | 0.068 | zgc:92518 | -0.015 |
| tbx20 | 0.067 | hhatla | -0.015 |
| drd7 | 0.067 | eif4ebp3l | -0.015 |
| fam131a | 0.067 | eif4ebp1 | -0.015 |
| si:ch211-191d15.2 | 0.065 | jph1b | -0.015 |
| crestin | 0.064 | nip7 | -0.015 |
| CR759863.2 | 0.064 | frem3 | -0.015 |