si:dkey-152b24.7
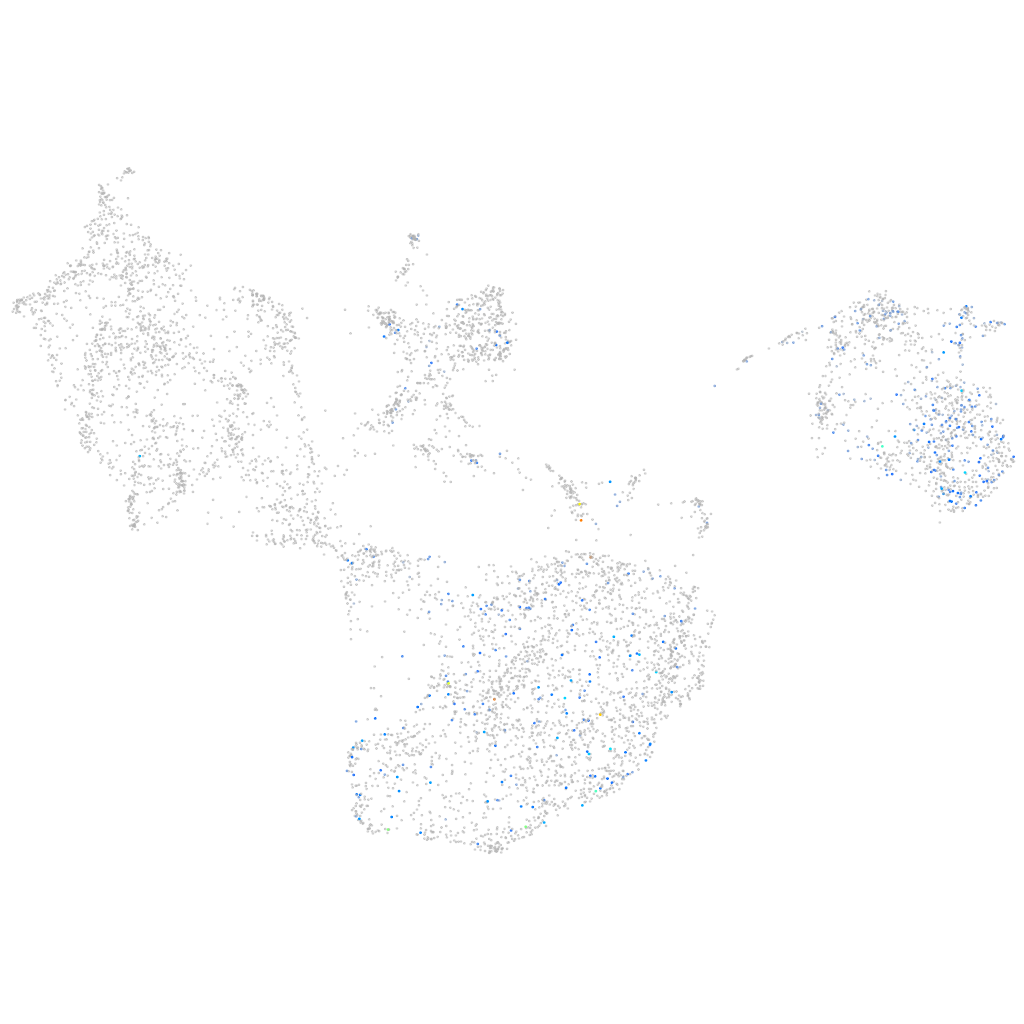
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line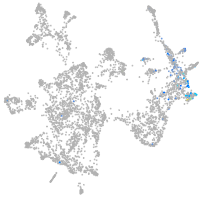 |
epidermis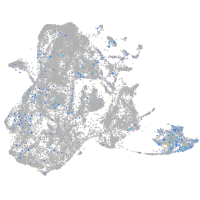 |
periderm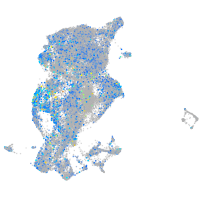 |
fin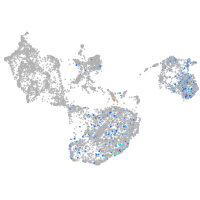 |
ionocytes / mucous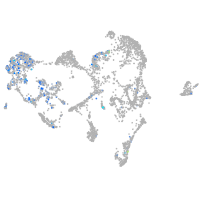 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnni4b.1 | 0.213 | c1qtnf5 | -0.126 |
| epcam | 0.180 | bhmt | -0.121 |
| cd9b | 0.178 | fbln1 | -0.116 |
| anxa1a | 0.171 | hgd | -0.113 |
| gabrg1 | 0.169 | rplp2l | -0.110 |
| pnp5a | 0.168 | rplp1 | -0.106 |
| si:dkey-248g15.3 | 0.168 | cd82a | -0.104 |
| si:ch211-157c3.4 | 0.161 | ttc36 | -0.103 |
| cldni | 0.161 | tmem119b | -0.103 |
| si:ch211-165d12.4 | 0.160 | twist1a | -0.102 |
| si:dkey-19b23.12 | 0.159 | pcbd1 | -0.102 |
| scpp8 | 0.158 | si:dkey-151g10.6 | -0.100 |
| zp3 | 0.157 | dcn | -0.100 |
| tmsb1 | 0.156 | prrx1a | -0.099 |
| epgn | 0.156 | qdpra | -0.098 |
| cldn23a | 0.156 | pmp22a | -0.098 |
| vamp8 | 0.156 | cfl1 | -0.098 |
| spint2 | 0.156 | ptx3a | -0.098 |
| cfl1l | 0.155 | rps28 | -0.098 |
| wu:fb18f06 | 0.155 | hpdb | -0.097 |
| s100v2 | 0.155 | prrx1b | -0.096 |
| lye | 0.154 | rps12 | -0.095 |
| cyt1 | 0.153 | gpc1a | -0.094 |
| zgc:111983 | 0.152 | rps14 | -0.094 |
| s100a10b | 0.151 | ntd5 | -0.094 |
| icn | 0.151 | mfap2 | -0.093 |
| stx11b.1 | 0.150 | rpl37 | -0.093 |
| zgc:174938 | 0.150 | reck | -0.093 |
| cldne | 0.149 | rps24 | -0.091 |
| evpla | 0.149 | marcksl1a | -0.089 |
| krt4 | 0.149 | fah | -0.088 |
| f11r.1 | 0.149 | hoxa13b | -0.088 |
| si:ch73-204p21.2 | 0.149 | col5a1 | -0.088 |
| pycard | 0.148 | rpl39 | -0.087 |
| tmem45b | 0.148 | slc16a10 | -0.086 |