si:dkey-150k17.3
ZFIN 
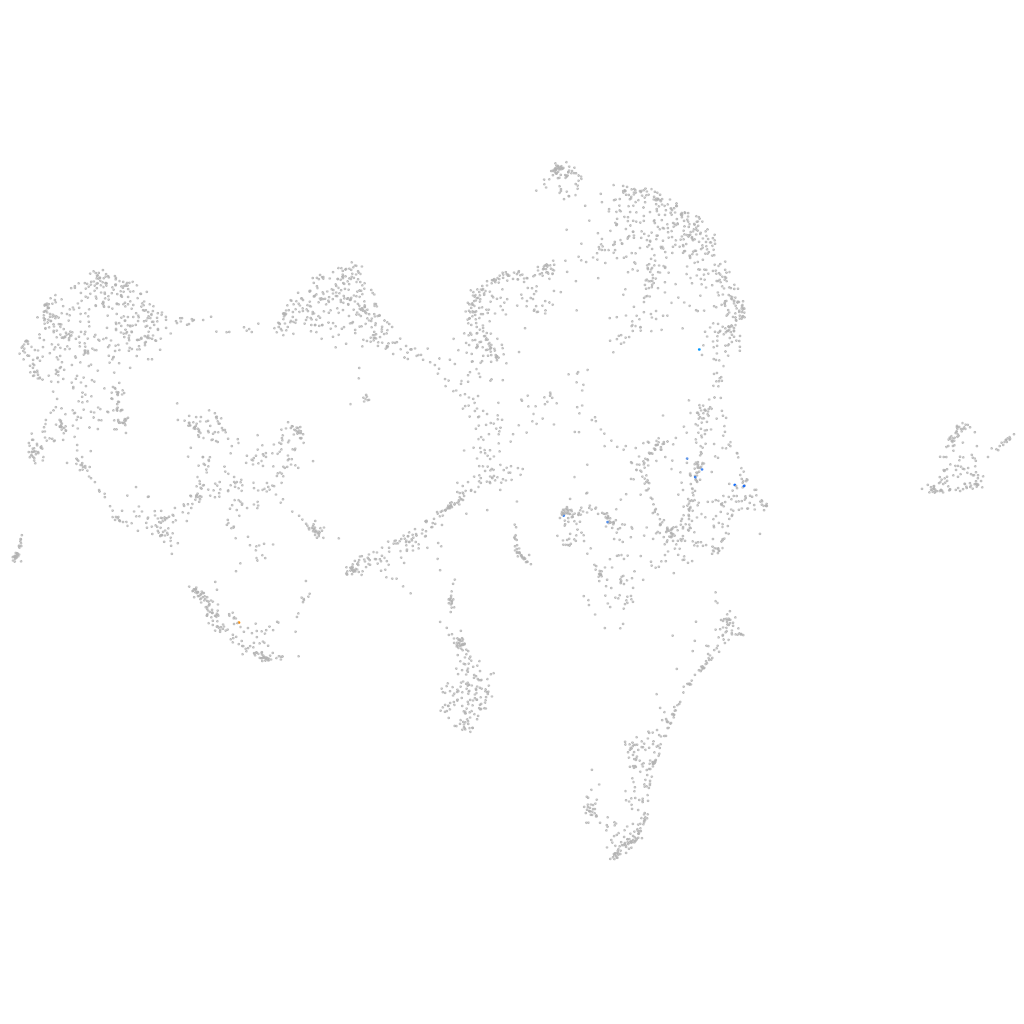
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:171759 | 0.611 | rps27a | -0.088 |
| hic1 | 0.572 | rpl34 | -0.065 |
| LOC110440146 | 0.528 | rps8a | -0.062 |
| smoc1 | 0.487 | rps4x | -0.056 |
| cilp | 0.478 | rpl9 | -0.055 |
| XLOC-026848 | 0.404 | nme2b.1 | -0.044 |
| odf2a | 0.382 | ahnak | -0.041 |
| spon2b | 0.381 | rps17 | -0.041 |
| ARMC6 | 0.345 | ier2b | -0.041 |
| CU571074.1 | 0.333 | cox6a1 | -0.040 |
| en1b | 0.328 | atp5meb | -0.039 |
| ccni2 | 0.328 | prr13 | -0.039 |
| ache | 0.321 | zgc:162730 | -0.039 |
| thbs4b | 0.310 | cfl1l | -0.038 |
| gxylt2 | 0.304 | btf3 | -0.038 |
| ghrb | 0.293 | ndufa6 | -0.037 |
| si:dkey-121j17.6 | 0.291 | tob1b | -0.037 |
| tmem169a | 0.286 | junba | -0.037 |
| acot7 | 0.278 | eef1da | -0.036 |
| nrip2 | 0.275 | zgc:110333 | -0.036 |
| wdfy4 | 0.257 | ybx1 | -0.035 |
| vwde | 0.247 | cd63 | -0.034 |
| yme1l1b | 0.245 | jun | -0.033 |
| si:rp71-1k13.6 | 0.224 | epcam | -0.033 |
| tead1a | 0.217 | ppdpfb | -0.033 |
| sort1a | 0.217 | mt-nd5 | -0.033 |
| CR790366.1 | 0.213 | romo1 | -0.033 |
| dzip1 | 0.209 | cox5aa | -0.033 |
| si:ch211-126c2.4 | 0.206 | soul2 | -0.033 |
| epg5 | 0.206 | malb | -0.033 |
| fgfr4 | 0.200 | zgc:153284 | -0.032 |
| il17rd | 0.192 | gstp1 | -0.032 |
| tmem248 | 0.190 | sod1 | -0.032 |
| phldb1b | 0.187 | hopx | -0.032 |
| rngtt | 0.187 | tmem258 | -0.032 |