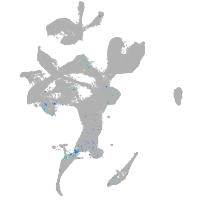
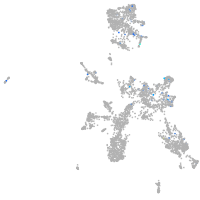
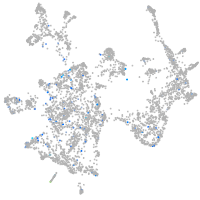
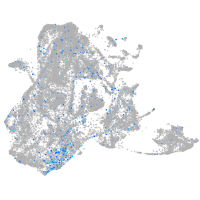
si:dkey-126g1.9
ZFIN 
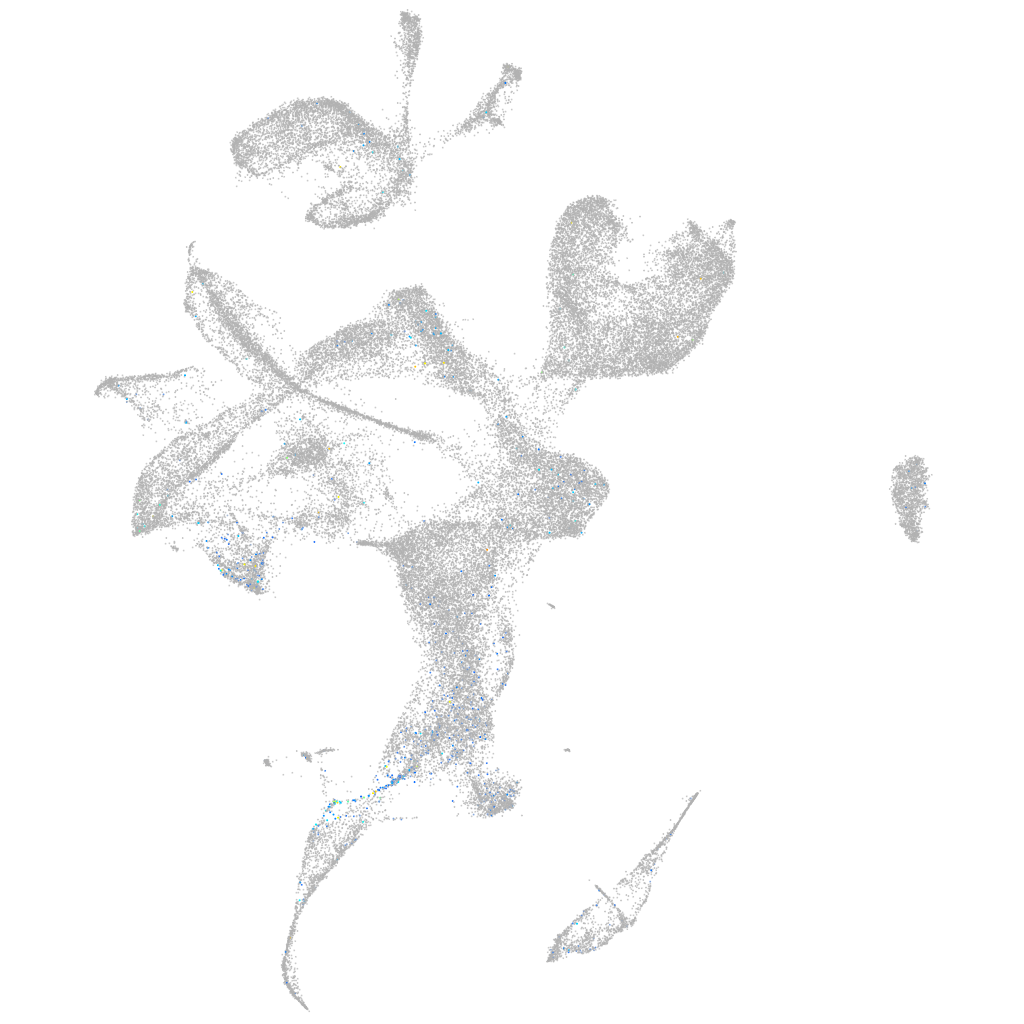
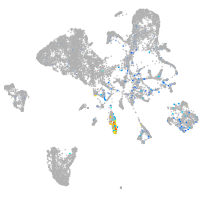
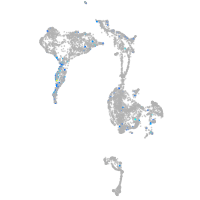
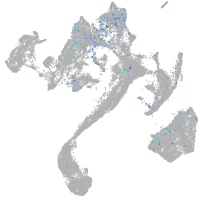
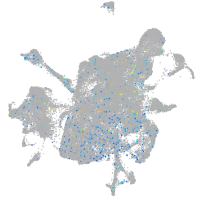
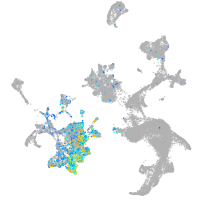
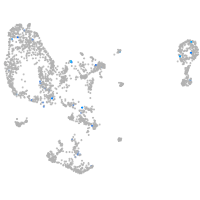
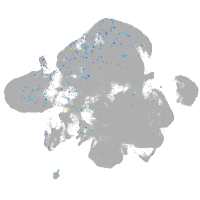
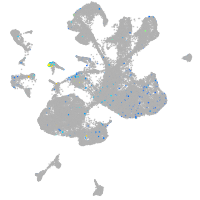
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| c1qtnf12 | 0.116 | gpm6aa | -0.049 |
| cdon | 0.111 | gpm6ab | -0.046 |
| pappa2 | 0.106 | ptmab | -0.045 |
| kcnk2a | 0.103 | ckbb | -0.044 |
| col15a1b | 0.102 | nova2 | -0.039 |
| fbln1 | 0.101 | marcksl1b | -0.039 |
| cldn19 | 0.096 | ndrg4 | -0.039 |
| zgc:153311 | 0.095 | si:ch73-1a9.3 | -0.038 |
| fabp11a | 0.095 | hmgn6 | -0.037 |
| cldn7a | 0.093 | hmgb1a | -0.035 |
| ggctb | 0.092 | cspg5a | -0.035 |
| nanos1 | 0.087 | vsx1 | -0.034 |
| wfdc1 | 0.087 | snap25b | -0.033 |
| CABZ01074363.1 | 0.086 | hnrnpa0a | -0.033 |
| ldlrap1a | 0.084 | gng13b | -0.032 |
| sfrp2 | 0.083 | scrt2 | -0.031 |
| mfap2 | 0.083 | rorb | -0.031 |
| col9a1b | 0.077 | rnasekb | -0.030 |
| ccdc3a | 0.076 | atp6v0cb | -0.030 |
| sytl2b | 0.074 | neurod4 | -0.030 |
| tpm4a | 0.074 | si:dkey-276j7.1 | -0.030 |
| trim35-10 | 0.074 | foxg1b | -0.030 |
| col9a2 | 0.074 | rtn1a | -0.029 |
| fstl1b | 0.074 | epb41a | -0.029 |
| kcnj1a.3 | 0.073 | calb2b | -0.029 |
| LOC101882770 | 0.073 | atp1b2a | -0.029 |
| cx43 | 0.072 | mdkb | -0.028 |
| sparc | 0.072 | ppp1r14c | -0.028 |
| CU927890.1 | 0.071 | pcbp3 | -0.028 |
| CU074421.1 | 0.070 | tuba1c | -0.028 |
| avpr2ab | 0.069 | stmn1b | -0.028 |
| clec19a | 0.069 | anp32e | -0.028 |
| pcolce2b | 0.069 | gng3 | -0.028 |
| pecam1 | 0.068 | gnb3a | -0.028 |
| gas1a | 0.068 | atp1a3a | -0.027 |