si:dkey-119f1.1
ZFIN 
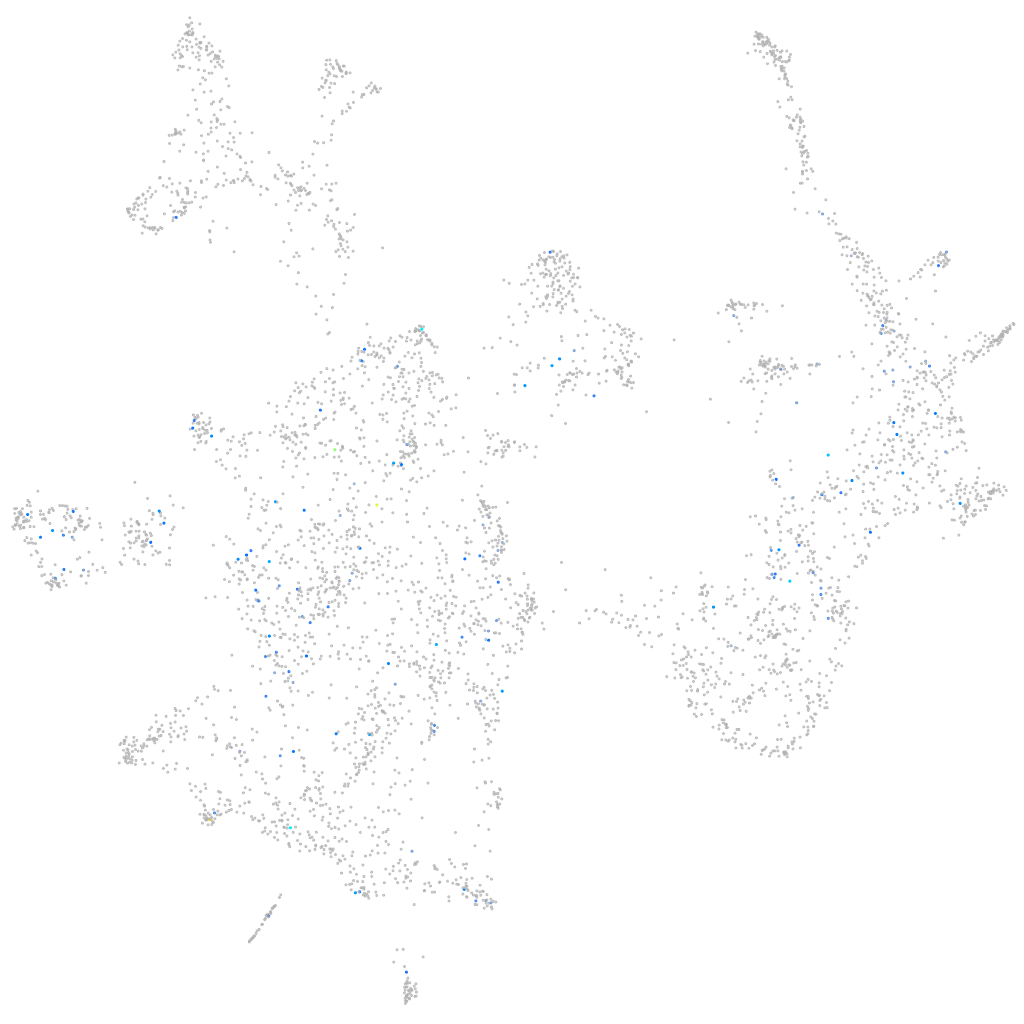
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc35a3b | 0.196 | calm1b | -0.059 |
| LO018568.2 | 0.188 | otofb | -0.059 |
| si:dkey-28d5.11 | 0.186 | gapdhs | -0.058 |
| si:ch73-252i11.1 | 0.181 | rnasekb | -0.058 |
| slc12a10.3 | 0.180 | nptna | -0.058 |
| il21r.1 | 0.173 | calml4a | -0.057 |
| CU457778.1 | 0.167 | eno1a | -0.057 |
| znfl1g | 0.163 | atp1a3b | -0.055 |
| pcna | 0.159 | cd164l2 | -0.055 |
| slbp | 0.154 | gpx2 | -0.054 |
| dhfr | 0.153 | tspan13b | -0.054 |
| CT955996.1 | 0.149 | prr15la | -0.054 |
| LOC103910884 | 0.149 | osbpl1a | -0.054 |
| tbpl2 | 0.148 | gipc3 | -0.053 |
| chaf1a | 0.147 | CR925719.1 | -0.052 |
| rfc2 | 0.146 | enkur | -0.052 |
| nasp | 0.146 | pkig | -0.052 |
| psmc3ip | 0.146 | calm1a | -0.052 |
| BX470188.1 | 0.145 | s100t | -0.052 |
| nacad | 0.143 | lrrc73 | -0.052 |
| fen1 | 0.142 | emb | -0.052 |
| nup85 | 0.141 | pcsk5a | -0.051 |
| lig1 | 0.140 | atp2b1a | -0.051 |
| si:ch73-281i18.4 | 0.138 | slc17a8 | -0.051 |
| ing5b | 0.138 | cd37 | -0.050 |
| dut | 0.137 | si:ch73-15n15.3 | -0.050 |
| tfcp2l1 | 0.137 | atp1b2b | -0.050 |
| rpa3 | 0.136 | si:ch211-284k5.2 | -0.049 |
| mcm10 | 0.136 | camk2n1a | -0.049 |
| dck | 0.135 | zgc:162989 | -0.049 |
| chaf1b | 0.135 | gb:eh456644 | -0.049 |
| LO018629.1 | 0.135 | pou4f3 | -0.049 |
| LOC100330864 | 0.134 | myo6b | -0.049 |
| CABZ01005379.1 | 0.134 | syt14b | -0.049 |
| zgc:171604 | 0.133 | kif1aa | -0.049 |