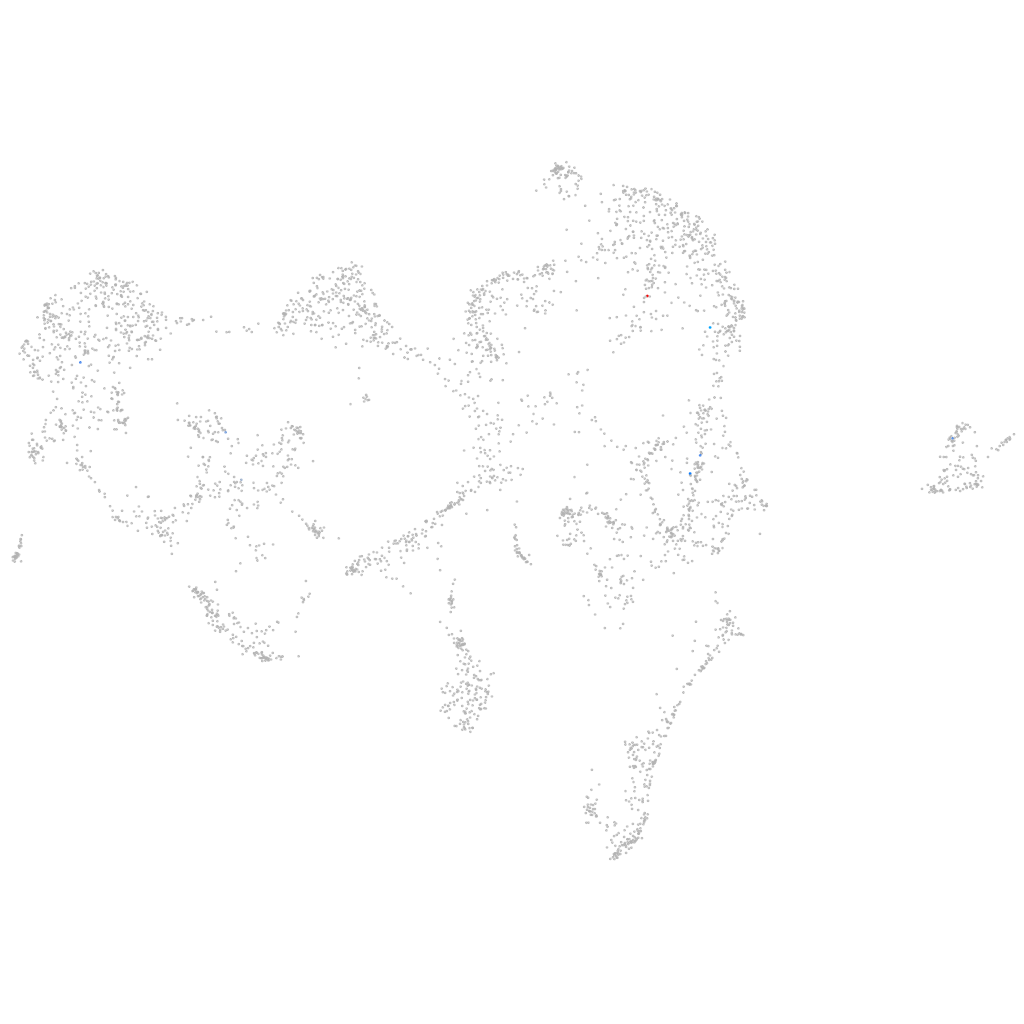
si:dkey-111e8.4
ZFIN 
Other cell groups


all cells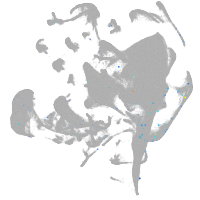 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 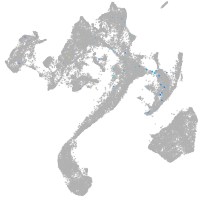 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin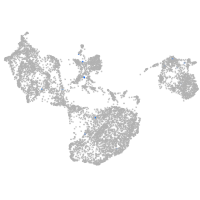 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dyrk4 | 0.657 | rplp0 | -0.095 |
| CABZ01048402.2 | 0.585 | rpl17 | -0.077 |
| gfm2 | 0.406 | rpl27 | -0.075 |
| actn3b | 0.380 | rpl8 | -0.074 |
| cdc42l2 | 0.342 | rps12 | -0.074 |
| myom1a | 0.342 | rpl24 | -0.074 |
| plppr3a | 0.324 | rpl30 | -0.073 |
| mysm1 | 0.315 | rpl14 | -0.072 |
| thap1 | 0.282 | rps4x | -0.068 |
| ccdc106a | 0.214 | hsp90ab1 | -0.066 |
| neb | 0.209 | rpl10a | -0.066 |
| si:ch211-122f10.4 | 0.207 | actb2 | -0.057 |
| itpkb | 0.201 | rps16 | -0.055 |
| si:zfos-2330d3.1 | 0.196 | btf3 | -0.053 |
| si:ch211-149l1.2 | 0.195 | cox4i1 | -0.049 |
| tmprss13b | 0.190 | cox8a | -0.048 |
| natd1 | 0.189 | rps17 | -0.048 |
| zgc:66484 | 0.186 | rps27.2 | -0.048 |
| thbs1b | 0.181 | mt-co1 | -0.046 |
| btbd2a | 0.180 | atp5f1b | -0.046 |
| tspan2b | 0.177 | eef1g | -0.044 |
| tmem43 | 0.176 | eef2b | -0.044 |
| txndc11 | 0.173 | atp5f1e | -0.044 |
| LO018176.1 | 0.172 | rps3 | -0.042 |
| tspy | 0.170 | ndufa4l | -0.041 |
| vamp4 | 0.164 | mt-nd1 | -0.041 |
| kmt5b | 0.164 | vdac2 | -0.040 |
| rab1aa | 0.154 | cox7c | -0.040 |
| naxe | 0.154 | atp5fa1 | -0.039 |
| p4ha1b | 0.149 | cox5aa | -0.039 |
| dop1b | 0.144 | fthl27 | -0.039 |
| cpsf3 | 0.142 | atp5meb | -0.037 |
| kmt2d | 0.139 | si:dkey-33i11.4 | -0.037 |
| si:dkey-1h24.2 | 0.137 | cox6c | -0.037 |
| slc26a2 | 0.135 | atp5f1d | -0.037 |