si:dkey-100n23.3
ZFIN 
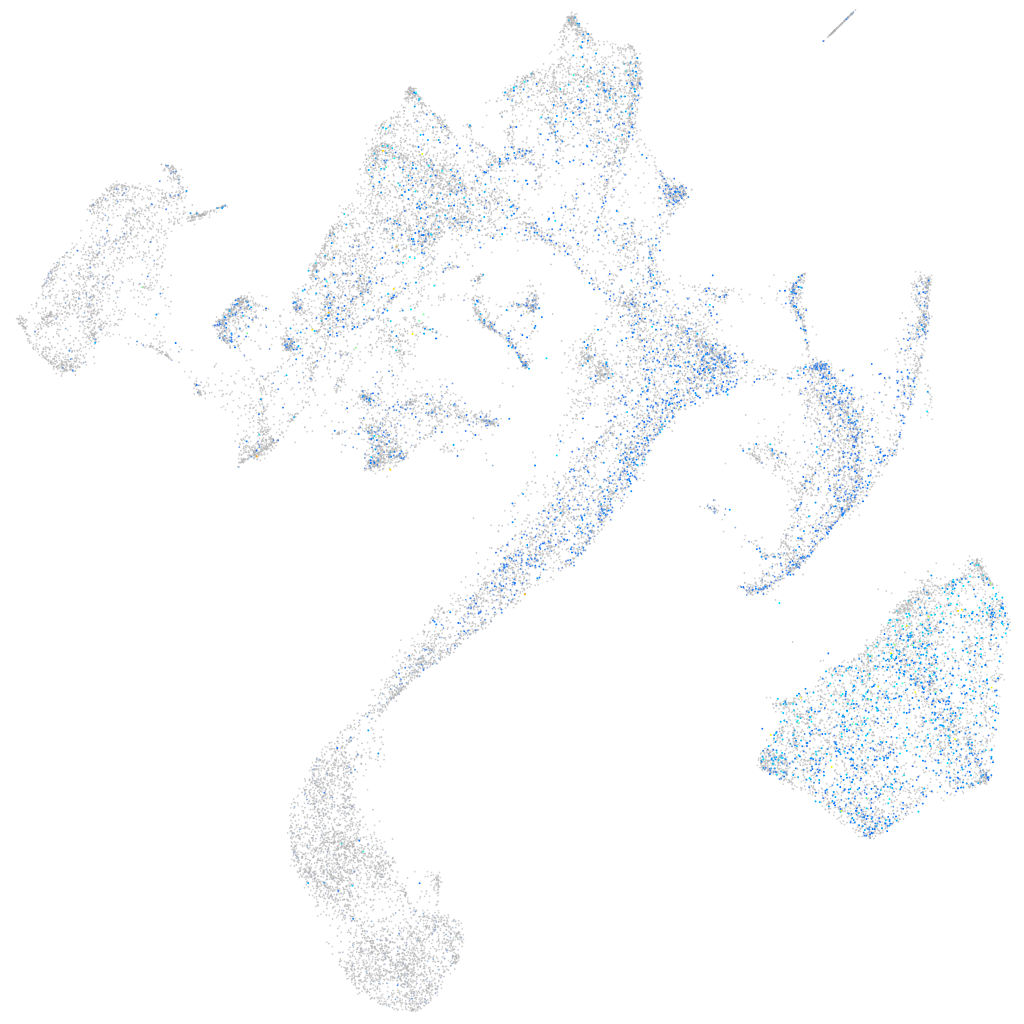
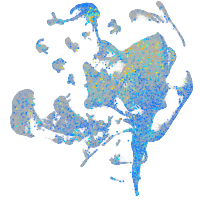
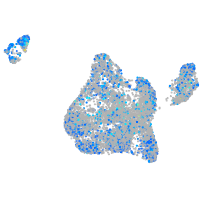
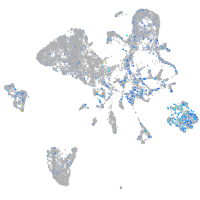
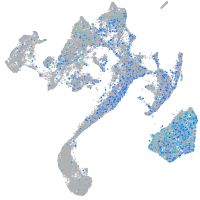
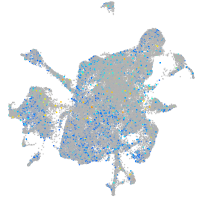
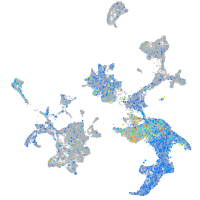
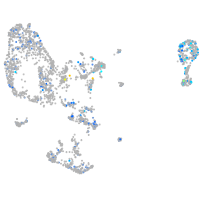
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ncl | 0.210 | actc1b | -0.170 |
| nucks1a | 0.207 | rpl37 | -0.163 |
| nop56 | 0.199 | ckma | -0.161 |
| anp32e | 0.199 | ak1 | -0.161 |
| fbl | 0.198 | ckmb | -0.159 |
| nop58 | 0.198 | atp2a1 | -0.153 |
| hmga1a | 0.198 | tnnc2 | -0.153 |
| syncrip | 0.198 | neb | -0.151 |
| hnrnpabb | 0.198 | aldoab | -0.150 |
| cirbpa | 0.196 | zgc:114188 | -0.149 |
| marcksb | 0.194 | eno3 | -0.147 |
| acin1a | 0.194 | ldb3b | -0.145 |
| khdrbs1a | 0.193 | actn3a | -0.144 |
| hnrnpub | 0.192 | si:ch73-367p23.2 | -0.143 |
| hnrnpa1b | 0.192 | acta1b | -0.143 |
| dkc1 | 0.191 | rps10 | -0.143 |
| npm1a | 0.190 | pvalb1 | -0.143 |
| seta | 0.190 | mylpfa | -0.143 |
| ilf3b | 0.189 | nme2b.2 | -0.142 |
| nop2 | 0.189 | actn3b | -0.142 |
| ppig | 0.188 | tpi1b | -0.141 |
| hnrnpaba | 0.188 | pvalb2 | -0.141 |
| rbm4.3 | 0.186 | tpma | -0.141 |
| hnrnpa0b | 0.186 | tmod4 | -0.141 |
| hnrnpa1a | 0.186 | gapdh | -0.141 |
| cx43.4 | 0.185 | pgam2 | -0.139 |
| ddx18 | 0.184 | smyd1a | -0.139 |
| setb | 0.184 | tmem38a | -0.138 |
| snrnp70 | 0.184 | mylz3 | -0.138 |
| bms1 | 0.183 | myom1a | -0.138 |
| snrpb | 0.183 | vdac3 | -0.138 |
| pabpc1a | 0.183 | si:ch211-266g18.10 | -0.137 |
| si:ch73-1a9.3 | 0.182 | ank1a | -0.137 |
| anp32a | 0.182 | eno1a | -0.137 |
| ebna1bp2 | 0.181 | rps17 | -0.137 |