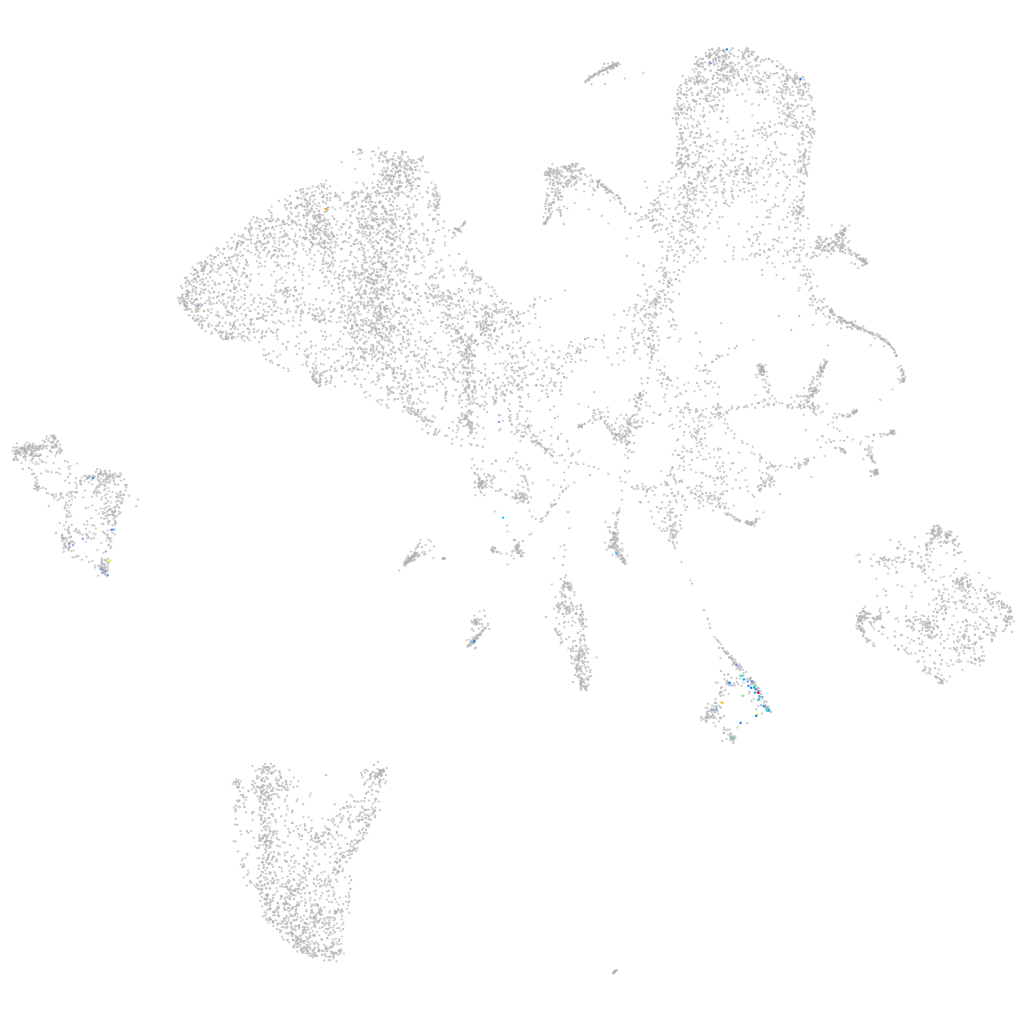
si:ch73-329n5.1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm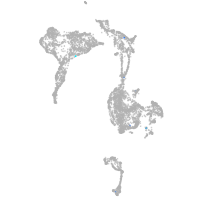 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory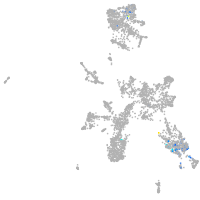 |
otic / lateral line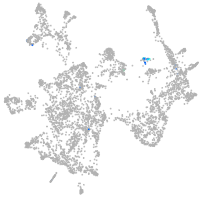 |
epidermis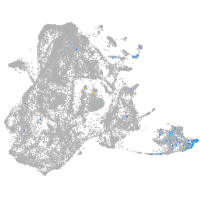 |
periderm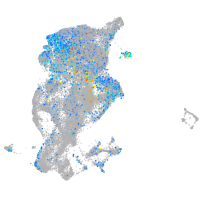 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-47f2.1 | 0.395 | cox6a1 | -0.059 |
| pnp4a | 0.373 | atp5mc3b | -0.059 |
| cd151 | 0.352 | gapdh | -0.059 |
| dnah11 | 0.328 | atp5l | -0.057 |
| si:dkey-30c15.2 | 0.296 | hspe1 | -0.053 |
| mxra8b | 0.289 | npm1a | -0.053 |
| tekt3 | 0.287 | aldh6a1 | -0.052 |
| CABZ01055365.1 | 0.285 | atp5mc1 | -0.052 |
| f3a | 0.283 | gatm | -0.051 |
| pmp22b | 0.278 | adh5 | -0.051 |
| amotl2b | 0.277 | si:dkey-16p21.8 | -0.051 |
| ctgfa | 0.276 | hspd1 | -0.050 |
| spns2 | 0.263 | aqp12 | -0.050 |
| hspa12b | 0.261 | mat1a | -0.050 |
| vat1l | 0.256 | ndufab1b | -0.049 |
| gna15.1 | 0.249 | gpx4a | -0.049 |
| tmem154 | 0.248 | pklr | -0.047 |
| capn2a | 0.245 | gamt | -0.047 |
| cygb1 | 0.244 | tpi1b | -0.047 |
| sim1b | 0.240 | cox7c | -0.047 |
| cav1 | 0.238 | bhmt | -0.047 |
| krt94 | 0.238 | pa2g4a | -0.047 |
| spaca4l | 0.237 | mt-atp6 | -0.046 |
| sftpba | 0.237 | phb2b | -0.046 |
| cabp2b | 0.234 | slc25a20 | -0.046 |
| si:dkey-126g1.7 | 0.233 | atp5fa1 | -0.046 |
| anxa3b | 0.229 | ahcy | -0.046 |
| sparc | 0.226 | mdh1aa | -0.046 |
| ccl20b | 0.221 | gcshb | -0.046 |
| adcy2b | 0.219 | prdx4 | -0.046 |
| abca12 | 0.217 | lgals2b | -0.046 |
| lrata | 0.216 | si:ch1073-325m22.2 | -0.046 |
| slbp2 | 0.215 | chchd10 | -0.045 |
| col4a6 | 0.214 | nipsnap3a | -0.045 |
| col4a5 | 0.214 | pcbd1 | -0.045 |