si:ch73-288o11.4
ZFIN 
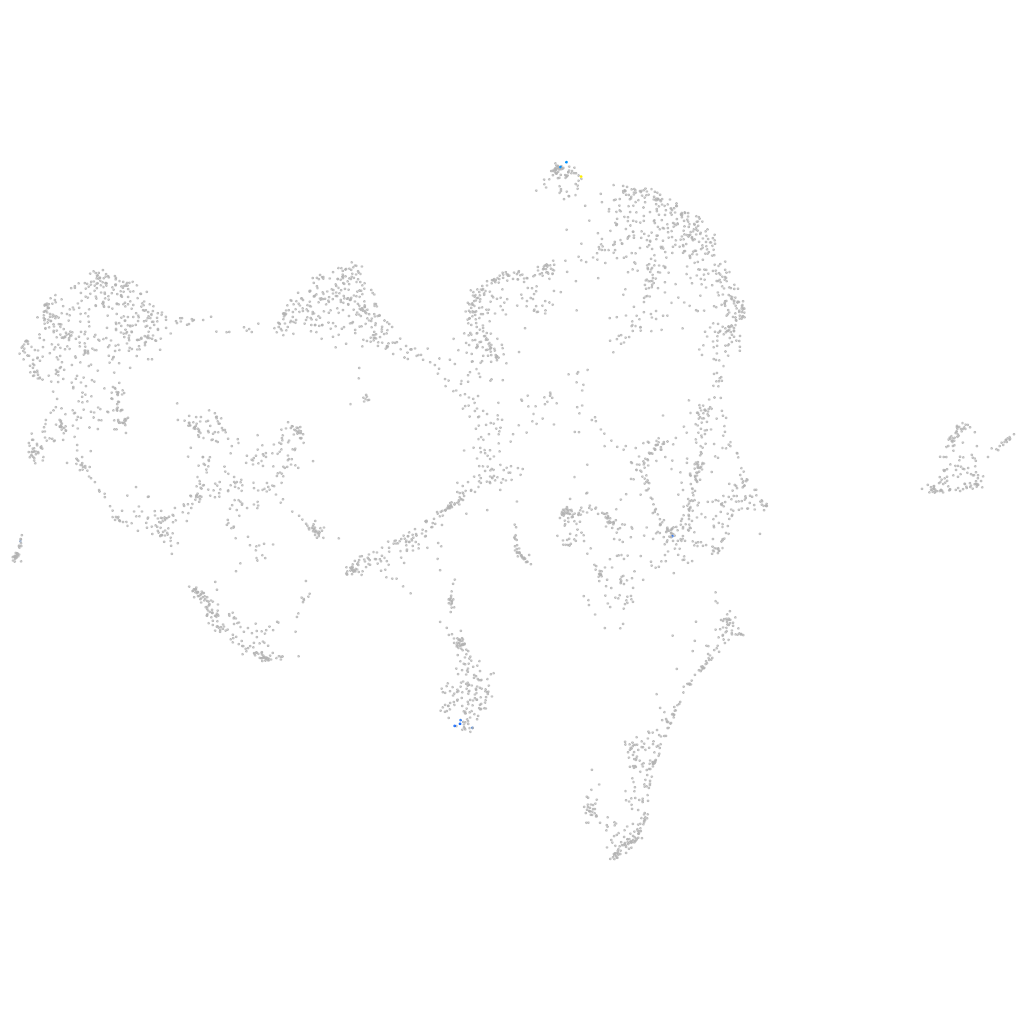
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 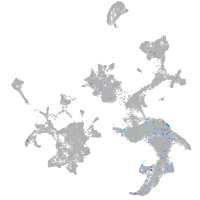 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous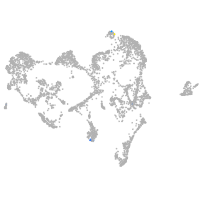 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-252p3.1 | 0.526 | rpl19 | -0.065 |
| sncaip | 0.523 | rps26 | -0.051 |
| gnat1 | 0.375 | rps16 | -0.050 |
| ly6m5 | 0.371 | rpl18a | -0.045 |
| si:dkey-234i14.3 | 0.335 | eef1g | -0.042 |
| CABZ01067151.2 | 0.312 | pabpc1a | -0.041 |
| AL928650.1 | 0.295 | eif3d | -0.038 |
| CU019662.1 | 0.286 | btg2 | -0.037 |
| tanc1b | 0.284 | rpl3 | -0.037 |
| lrrc4bb | 0.282 | mt-nd3 | -0.036 |
| foxe1 | 0.278 | calm2b | -0.035 |
| LOC110440119 | 0.276 | atp5fa1 | -0.034 |
| kcnj13 | 0.275 | rps27.2 | -0.034 |
| fgg | 0.259 | ndufa3 | -0.034 |
| rcvrn3 | 0.255 | mt-nd4 | -0.034 |
| kirrel1a | 0.250 | actb1 | -0.033 |
| sec22c | 0.242 | si:dkey-16p21.8 | -0.032 |
| cipca | 0.232 | rnf10 | -0.032 |
| pgam2 | 0.224 | ndufa1 | -0.032 |
| rnf41l | 0.215 | gng5 | -0.032 |
| mfsd12a | 0.207 | pnrc2 | -0.031 |
| si:dkey-19a16.7 | 0.202 | atrx | -0.031 |
| CABZ01083943.1 | 0.200 | arhgdia | -0.030 |
| zgc:194252 | 0.200 | rpl6 | -0.030 |
| tmem231 | 0.197 | ctnnb1 | -0.030 |
| mapk1 | 0.193 | mt-co1 | -0.030 |
| si:ch211-120g10.1 | 0.189 | sem1 | -0.030 |
| LOC110437709 | 0.188 | krtt1c19e | -0.030 |
| AL935183.1 | 0.188 | gadd45ba | -0.030 |
| ankfy1 | 0.184 | dusp2 | -0.030 |
| armt1 | 0.176 | emc2 | -0.029 |
| si:ch211-238g23.1 | 0.175 | sod2 | -0.029 |
| myom2a | 0.170 | tra2a | -0.029 |
| phf12a | 0.169 | eef1da | -0.029 |
| pargl | 0.169 | u2af2b | -0.029 |