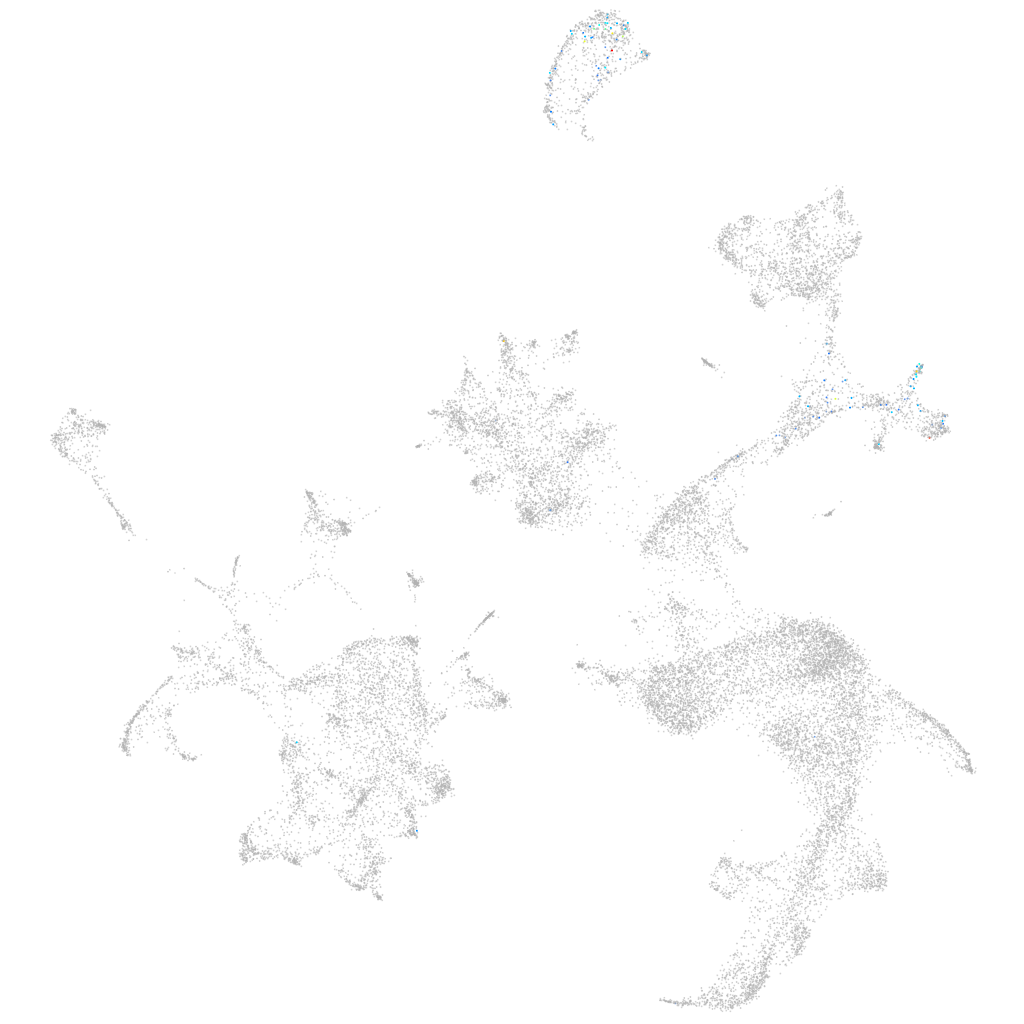
si:ch73-285p12.4
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1c2 | 0.171 | hbae3 | -0.058 |
| hspb7 | 0.171 | hbbe1.3 | -0.057 |
| sult1st3 | 0.157 | hbae1.1 | -0.056 |
| scinlb | 0.152 | hbae1.3 | -0.055 |
| aqp10a | 0.145 | hbbe2 | -0.053 |
| wasb | 0.140 | hbbe1.1 | -0.052 |
| nitr9 | 0.136 | cahz | -0.051 |
| prkcda | 0.135 | hbbe1.2 | -0.049 |
| il34 | 0.131 | fth1a | -0.048 |
| ponzr6 | 0.129 | blvrb | -0.048 |
| mmp13a | 0.128 | hemgn | -0.047 |
| LOC101882020 | 0.127 | zgc:56095 | -0.045 |
| coro1a | 0.127 | si:ch211-250g4.3 | -0.044 |
| rac2 | 0.126 | alas2 | -0.044 |
| LOC110440027 | 0.125 | slc4a1a | -0.042 |
| ccr9a | 0.124 | ctsla | -0.041 |
| dusp2 | 0.124 | creg1 | -0.041 |
| CR383669.1 | 0.123 | nt5c2l1 | -0.041 |
| cfbl | 0.123 | ap2m1a | -0.040 |
| zgc:136605 | 0.122 | lmo2 | -0.040 |
| si:ch211-67e16.3 | 0.121 | epb41b | -0.039 |
| LOC108185170 | 0.121 | zgc:163057 | -0.039 |
| ctss1 | 0.120 | tspo | -0.039 |
| chia.6 | 0.119 | si:ch211-207c6.2 | -0.038 |
| FP236331.1 | 0.118 | histh1l | -0.037 |
| si:ch73-248e21.5 | 0.118 | nmt1b | -0.036 |
| scpp8 | 0.117 | tmod4 | -0.035 |
| mmp9 | 0.117 | marcksl1b | -0.034 |
| npsn | 0.117 | aqp1a.1 | -0.033 |
| LOC110439930 | 0.117 | prdx2 | -0.033 |
| cpa5 | 0.116 | si:ch211-227m13.1 | -0.032 |
| siglec15l | 0.116 | hbae5 | -0.032 |
| si:dkey-262k9.4 | 0.114 | sptb | -0.031 |
| BX510306.1 | 0.114 | clic2 | -0.031 |
| cd7al | 0.114 | hbbe3 | -0.031 |