si:ch73-266f23.3
ZFIN 
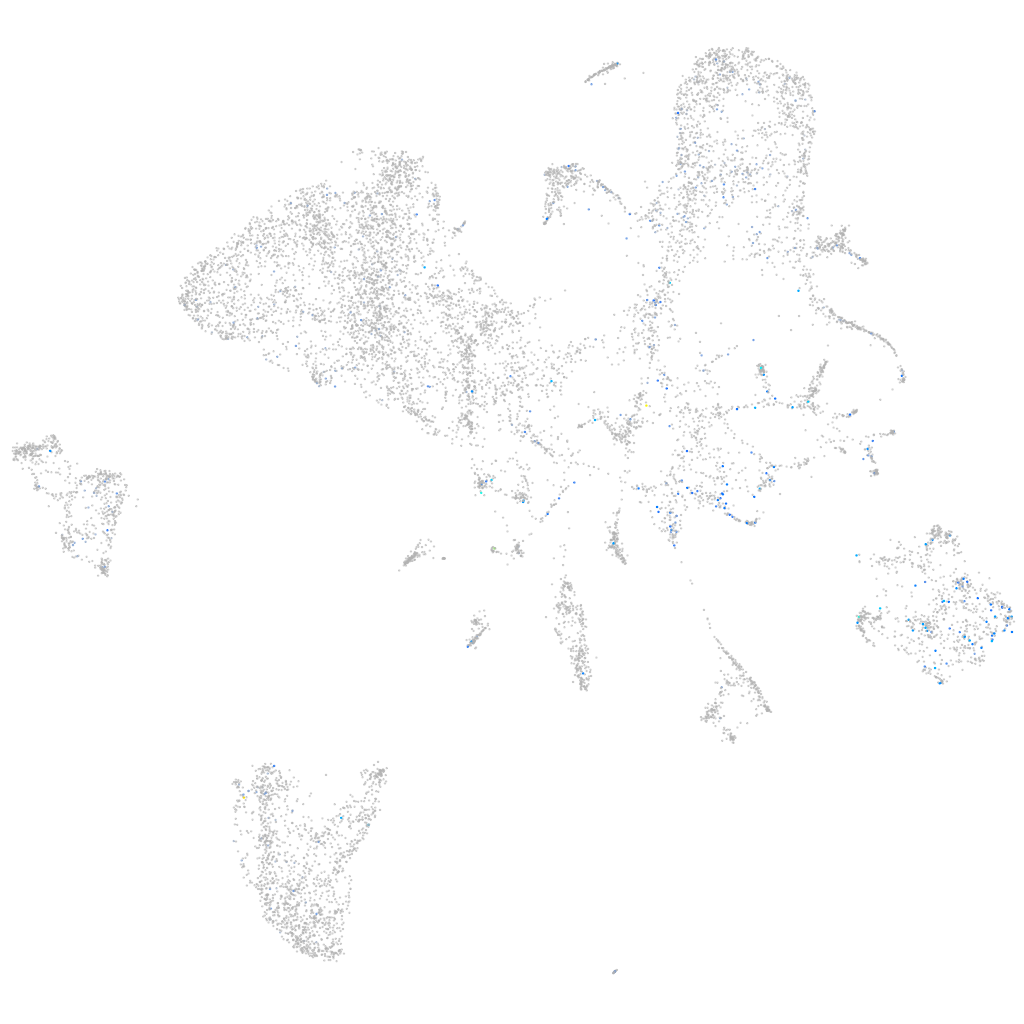
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-15b23.3 | 0.178 | gapdh | -0.092 |
| marcksb | 0.143 | gamt | -0.091 |
| hnrnpaba | 0.140 | gatm | -0.085 |
| seta | 0.140 | bhmt | -0.085 |
| ddx39aa | 0.137 | ahcy | -0.084 |
| khdrbs1a | 0.137 | apoa2 | -0.080 |
| hmga1a | 0.136 | apoa1b | -0.079 |
| hdac1 | 0.136 | agxtb | -0.078 |
| syncrip | 0.135 | pnp4b | -0.076 |
| cbx1a | 0.134 | mat1a | -0.076 |
| ddx39ab | 0.134 | fbp1b | -0.075 |
| cbx3a | 0.133 | rbp4 | -0.073 |
| hnrnpabb | 0.133 | eno3 | -0.073 |
| hnrnpa1b | 0.132 | gpx4a | -0.073 |
| smarca4a | 0.132 | zgc:123103 | -0.073 |
| taf15 | 0.131 | afp4 | -0.073 |
| nucks1a | 0.131 | apoa4b.1 | -0.072 |
| h3f3d | 0.130 | fabp10a | -0.072 |
| si:ch211-222l21.1 | 0.129 | rpl37 | -0.072 |
| hnrnph1l | 0.129 | tfa | -0.071 |
| rbm4.2 | 0.128 | cebpd | -0.071 |
| smarcd1 | 0.128 | agxta | -0.071 |
| si:ch211-152c2.3 | 0.127 | ckba | -0.070 |
| si:ch73-281n10.2 | 0.127 | serpina1 | -0.070 |
| CT990561.2 | 0.127 | zgc:92744 | -0.070 |
| snrpb | 0.126 | rbp2b | -0.070 |
| hnrnpa0b | 0.126 | grhprb | -0.070 |
| ilf3b | 0.126 | apom | -0.069 |
| si:ch211-288g17.3 | 0.126 | serpina1l | -0.069 |
| rbm4.3 | 0.126 | LOC110437731 | -0.069 |
| hmgb2b | 0.126 | pklr | -0.069 |
| setb | 0.125 | hao1 | -0.069 |
| hmgb2a | 0.125 | fgb | -0.068 |
| cirbpb | 0.124 | ttr | -0.068 |
| sap18 | 0.124 | pgm1 | -0.068 |