si:ch73-237c6.1
ZFIN 
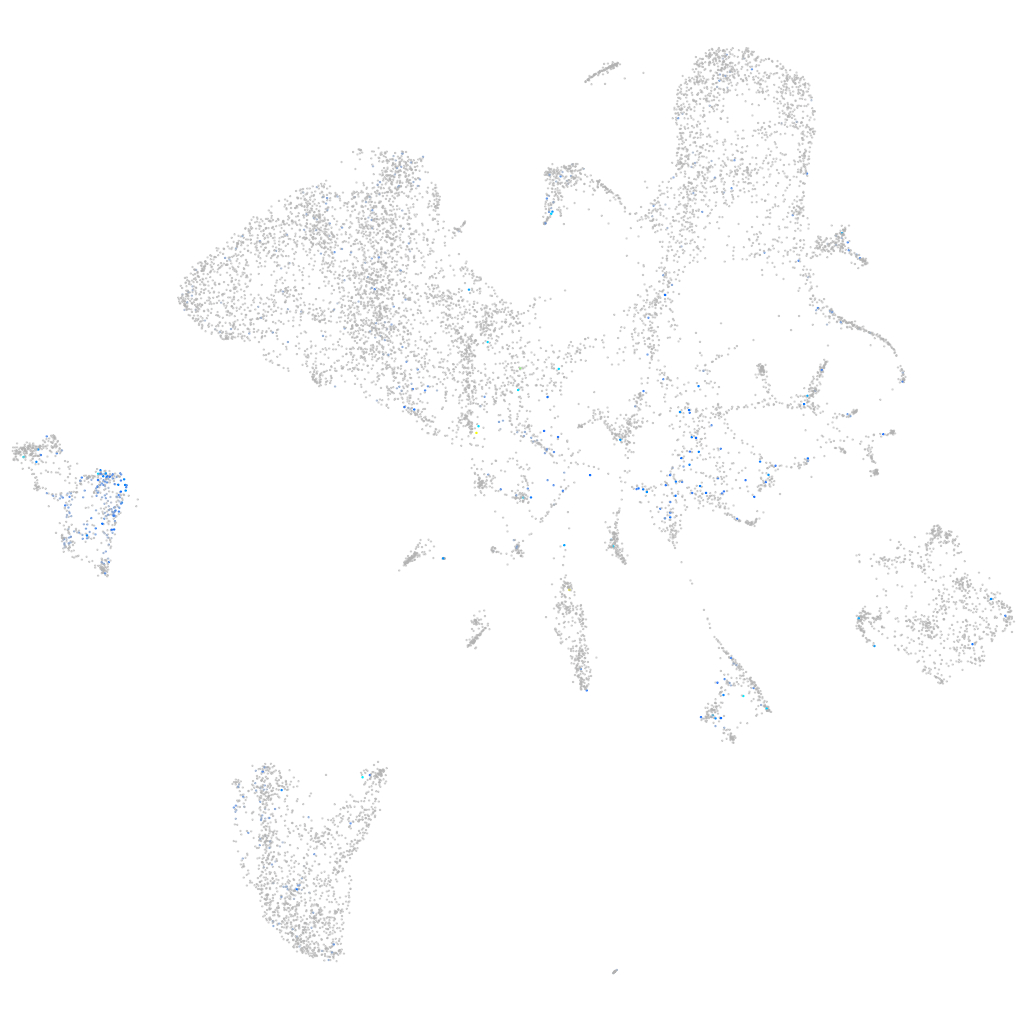
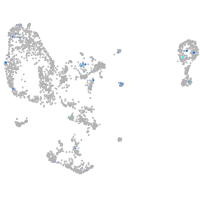
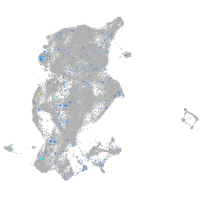
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:busm1-71b9.3 | 0.244 | gapdh | -0.114 |
| igfbp7 | 0.239 | apoc2 | -0.093 |
| cldn1 | 0.231 | apoa4b.1 | -0.093 |
| cldni | 0.219 | gpx4a | -0.090 |
| si:rp71-77l1.1 | 0.203 | scp2a | -0.089 |
| ca6 | 0.203 | afp4 | -0.086 |
| lamb4 | 0.196 | gatm | -0.085 |
| tp63 | 0.193 | gamt | -0.085 |
| si:dkey-102c8.3 | 0.193 | fbp1b | -0.082 |
| col4a5 | 0.186 | lgals2b | -0.082 |
| epgn | 0.185 | apoa1b | -0.078 |
| anxa2a | 0.182 | pklr | -0.078 |
| col4a6 | 0.181 | glud1b | -0.076 |
| zgc:101810 | 0.179 | suclg1 | -0.075 |
| cxl34b.11 | 0.178 | dhrs9 | -0.075 |
| caspb | 0.178 | sult2st2 | -0.075 |
| anos1a | 0.177 | msrb2 | -0.075 |
| shhb | 0.176 | apoc1 | -0.074 |
| LOC101885053 | 0.175 | rdh1 | -0.074 |
| ptgs2a | 0.174 | apobb.1 | -0.073 |
| myoc | 0.170 | gpd1b | -0.073 |
| zgc:153911 | 0.170 | bhmt | -0.072 |
| kitlgb | 0.169 | gstt1a | -0.072 |
| sparc | 0.169 | gcshb | -0.071 |
| fat2 | 0.168 | pnp4b | -0.070 |
| col5a2a | 0.166 | dap | -0.069 |
| igfbp5b | 0.166 | slco1d1 | -0.069 |
| gpa33a | 0.165 | cox7a1 | -0.069 |
| cfl1l | 0.165 | tpi1b | -0.068 |
| si:ch1073-406l10.2 | 0.164 | slc37a4a | -0.068 |
| foxe1 | 0.163 | tdo2a | -0.068 |
| kitlga | 0.162 | acmsd | -0.068 |
| CR450727.1 | 0.161 | apoa2 | -0.067 |
| ccl27a | 0.161 | etnppl | -0.067 |
| pfn1 | 0.157 | abat | -0.066 |