si:ch73-147o17.1
ZFIN 
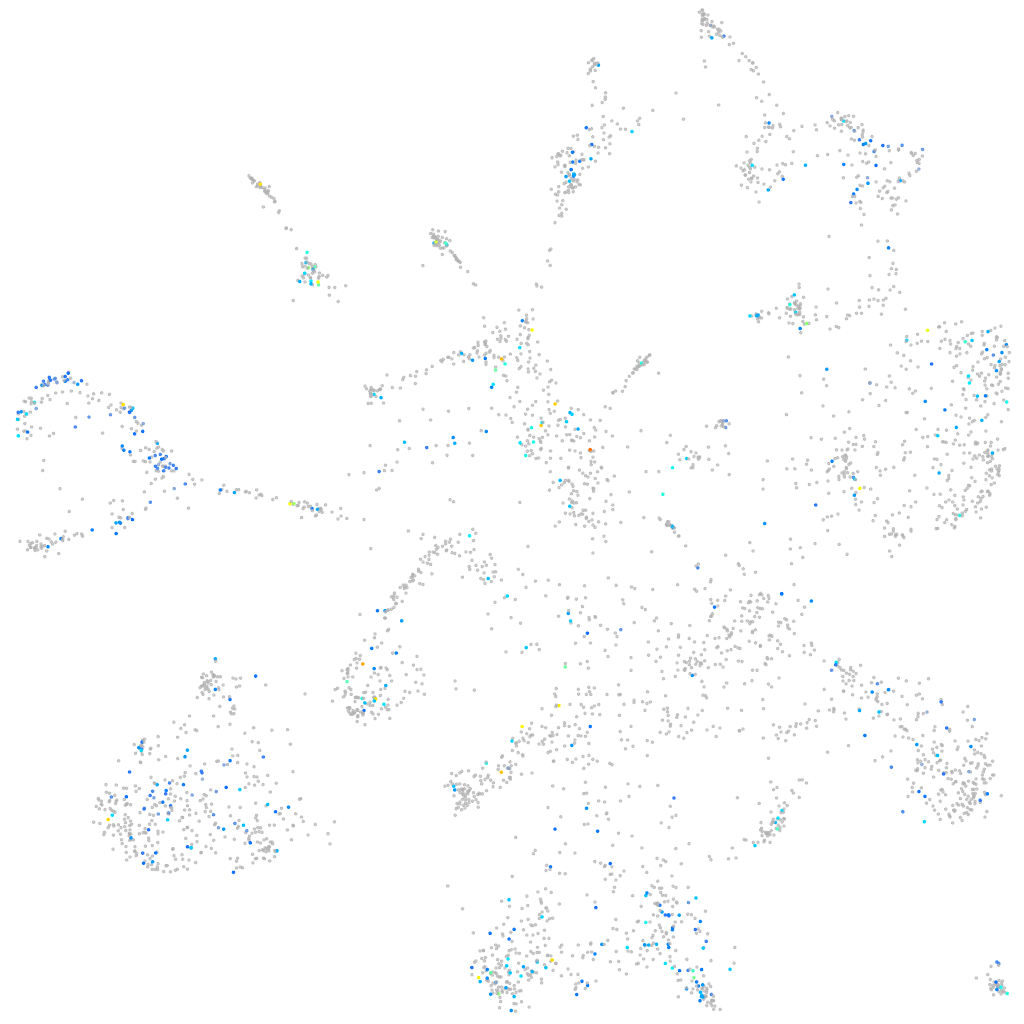
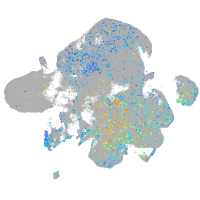
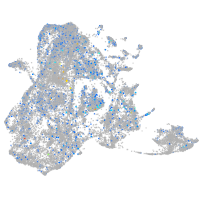
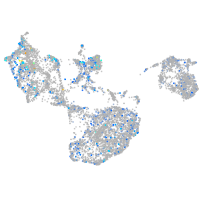
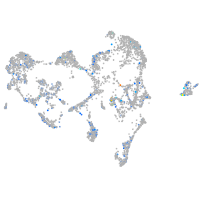
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rbm24a | 0.144 | hmga1a | -0.073 |
| acta2 | 0.125 | hmgb2a | -0.071 |
| tpm2 | 0.122 | seta | -0.071 |
| csrp1b | 0.121 | si:ch73-1a9.3 | -0.070 |
| tpm1 | 0.120 | stmn1a | -0.068 |
| pfn1 | 0.115 | si:ch211-222l21.1 | -0.067 |
| ckba | 0.113 | hmgn2 | -0.063 |
| myl6 | 0.112 | smc1al | -0.062 |
| tagln | 0.111 | si:dkey-261h17.1 | -0.062 |
| myl9a | 0.111 | fbl | -0.061 |
| cald1b | 0.109 | nucks1a | -0.060 |
| cd4-1 | 0.106 | ppiaa | -0.059 |
| bves | 0.106 | hmgb2b | -0.059 |
| si:dkey-18a10.3 | 0.106 | apex1 | -0.058 |
| lxn | 0.105 | si:ch211-39i22.1 | -0.057 |
| zgc:153867 | 0.103 | ptmab | -0.056 |
| LOC103910227 | 0.101 | ppil1 | -0.055 |
| actc1a | 0.099 | ptpn13 | -0.055 |
| vdac1 | 0.098 | NC-002333.4 | -0.055 |
| ak1 | 0.097 | cdca7b | -0.055 |
| eno1a | 0.097 | tia1 | -0.054 |
| eno3 | 0.097 | nop56 | -0.054 |
| bsg | 0.096 | fgfr2 | -0.053 |
| ppp1cbl | 0.095 | zc3h7bb | -0.052 |
| tuba8l2 | 0.095 | si:ch73-281n10.2 | -0.052 |
| myh11a | 0.095 | snrpd1 | -0.052 |
| pkma | 0.095 | mcm6 | -0.052 |
| si:ch1073-190k2.1 | 0.094 | mcm2 | -0.051 |
| unm-sa821 | 0.094 | hells | -0.051 |
| atad3 | 0.094 | robo1 | -0.050 |
| pgk1 | 0.094 | tgif1 | -0.050 |
| def6a | 0.094 | nop58 | -0.049 |
| gapdhs | 0.094 | brd9 | -0.049 |
| gsna | 0.093 | BNC1 | -0.049 |
| pgam1a | 0.093 | wt1a | -0.048 |