si:ch73-111k22.2
ZFIN 
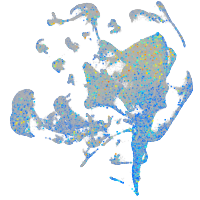
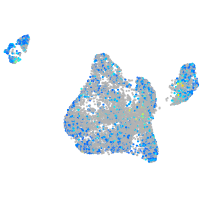
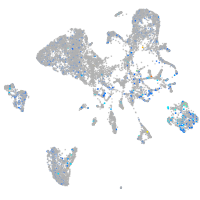
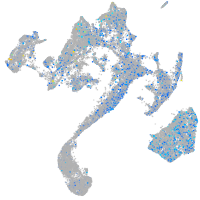
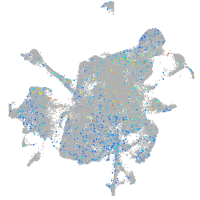
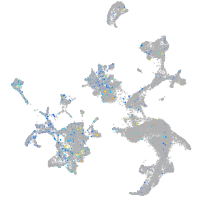
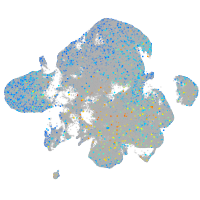
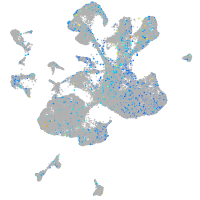
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.074 | fabp7a | -0.052 |
| myt1a | 0.072 | atp1a1b | -0.049 |
| tubb5 | 0.071 | glula | -0.038 |
| myt1b | 0.071 | slc1a2b | -0.037 |
| ebf2 | 0.069 | mdkb | -0.036 |
| dlb | 0.068 | ppap2d | -0.036 |
| tuba1a | 0.066 | zgc:165461 | -0.035 |
| tp53inp2 | 0.063 | atp1b4 | -0.035 |
| nhlh2 | 0.062 | efhd1 | -0.034 |
| fscn1a | 0.058 | pvalb1 | -0.034 |
| jpt1b | 0.056 | slc1a3b | -0.033 |
| hnrnpa0a | 0.055 | CU467822.1 | -0.032 |
| hnrnpa1a | 0.054 | pvalb2 | -0.032 |
| jagn1a | 0.053 | si:ch211-66e2.5 | -0.031 |
| hnrnpaba | 0.053 | dap1b | -0.031 |
| smarca4a | 0.053 | cx43 | -0.030 |
| si:ch211-222l21.1 | 0.052 | mdka | -0.030 |
| dnaja2b | 0.052 | ctrb1 | -0.030 |
| atrx | 0.052 | mt2 | -0.030 |
| tmeff1b | 0.052 | ptn | -0.030 |
| hmgb3a | 0.051 | cd63 | -0.030 |
| nucks1a | 0.051 | cox4i2 | -0.029 |
| neurod4 | 0.051 | hspb15 | -0.028 |
| syncrip | 0.051 | si:dkey-85k7.7 | -0.027 |
| ilf2 | 0.050 | mfge8a | -0.027 |
| hmgn6 | 0.050 | nck1a | -0.027 |
| sept5a | 0.050 | apoa1b | -0.027 |
| si:dkey-280e21.3 | 0.050 | apoa2 | -0.027 |
| zfhx3 | 0.050 | si:ch1073-303k11.2 | -0.027 |
| chd4a | 0.050 | prss59.2 | -0.027 |
| tuba1c | 0.050 | si:dkey-7j14.6 | -0.027 |
| si:dkey-276j7.1 | 0.050 | id1 | -0.027 |
| insm1a | 0.049 | slc3a2a | -0.027 |
| si:ch211-133n4.4 | 0.049 | slc6a11b | -0.026 |
| spsb4a | 0.049 | cebpd | -0.026 |