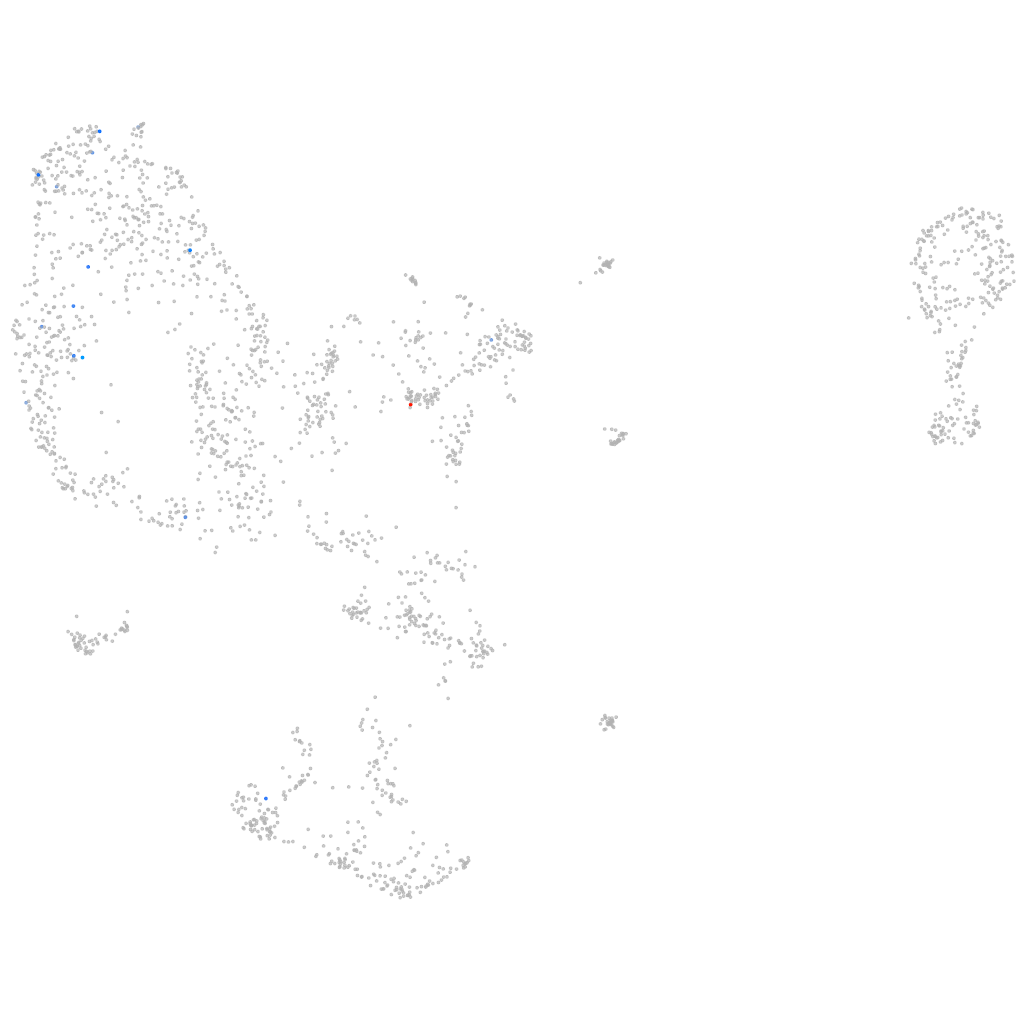
si:ch211-93f2.1 [Source:ZFIN;Acc:ZDB-GENE-041014-96]
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-284e13.14 | 0.818 | rplp2l | -0.153 |
| nr0b2a | 0.712 | rps20 | -0.133 |
| rbp1 | 0.685 | rpl26 | -0.118 |
| si:dkey-160o24.3 | 0.658 | rpl7 | -0.117 |
| bicdl2l | 0.654 | rpl12 | -0.113 |
| lingo1b | 0.598 | rpl18 | -0.110 |
| fstl5 | 0.558 | rps27.1 | -0.106 |
| frmd4bb | 0.539 | rps29 | -0.104 |
| cdh27 | 0.531 | rpl24 | -0.103 |
| bicc2 | 0.524 | rpl36 | -0.099 |
| CR848812.1 | 0.521 | rps11 | -0.094 |
| apba1a | 0.515 | rpl29 | -0.093 |
| vsig8b | 0.510 | rack1 | -0.092 |
| gpr37l1b | 0.506 | rpl34 | -0.091 |
| brf2 | 0.421 | rpl18a | -0.090 |
| ndst2b | 0.398 | rpl14 | -0.080 |
| XLOC-002129 | 0.397 | rpl23a | -0.079 |
| gdf10a | 0.389 | ptmab | -0.077 |
| camk1b | 0.372 | pabpc1a | -0.074 |
| itm2ca | 0.356 | rpl9 | -0.068 |
| tkta | 0.352 | hmgb2a | -0.063 |
| lmbrd2b | 0.350 | serbp1a | -0.058 |
| zgc:165515 | 0.347 | eef2b | -0.054 |
| myo9aa | 0.326 | hmga1a | -0.053 |
| stap2a | 0.322 | tubb4b | -0.053 |
| bmp7b | 0.317 | cirbpa | -0.052 |
| pgam1b | 0.316 | hmgn7 | -0.049 |
| si:ch1073-190k2.1 | 0.310 | hnrnpaba | -0.047 |
| hdac9b | 0.298 | setb | -0.046 |
| wu:fj16a03 | 0.297 | hnrnpa0b | -0.045 |
| l1cama | 0.294 | tma7 | -0.045 |
| gopc | 0.261 | hdac1 | -0.045 |
| TSTA3 (1 of many) | 0.260 | syncrip | -0.045 |
| mapk8a | 0.260 | sumo3a | -0.045 |
| and2 | 0.252 | rpl4 | -0.045 |