si:ch211-42i9.8
ZFIN 
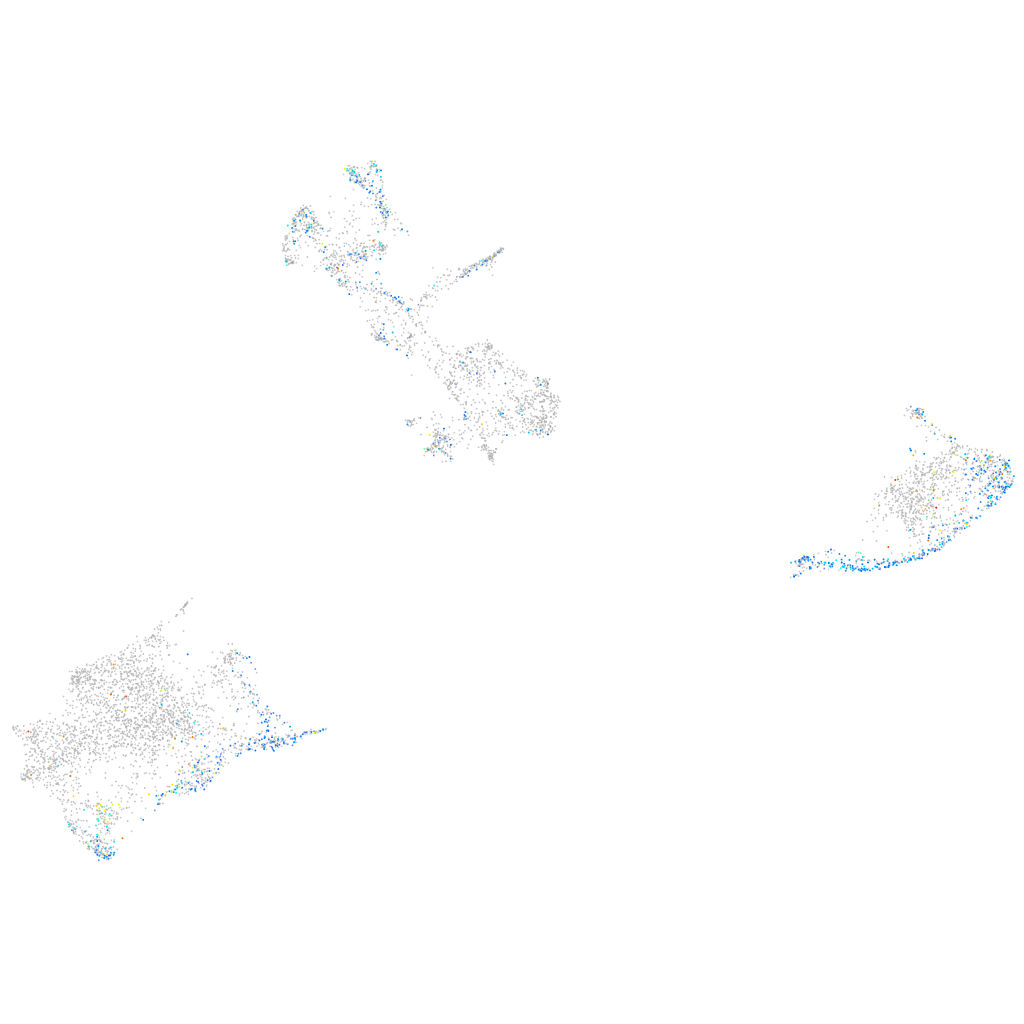
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.246 | CABZ01021592.1 | -0.125 |
| trpm1b | 0.209 | si:dkey-251i10.2 | -0.120 |
| si:zfos-943e10.1 | 0.199 | hbae3 | -0.103 |
| atp11a | 0.199 | uraha | -0.102 |
| bace2 | 0.191 | hbbe1.3 | -0.102 |
| tfap2e | 0.185 | ptmaa | -0.088 |
| mitfa | 0.184 | si:ch211-222l21.1 | -0.085 |
| tyr | 0.183 | tuba1c | -0.084 |
| slc45a2 | 0.181 | elavl3 | -0.080 |
| lamp1a | 0.180 | hbae1.1 | -0.080 |
| pim1 | 0.179 | gpm6aa | -0.079 |
| psap | 0.177 | pvalb2 | -0.077 |
| sdc4 | 0.175 | stmn1b | -0.077 |
| ctsc | 0.174 | hbbe1.1 | -0.076 |
| BX571715.1 | 0.171 | tmsb | -0.074 |
| rabl6b | 0.171 | nova2 | -0.073 |
| ctsba | 0.171 | actc1b | -0.071 |
| slc3a2a | 0.170 | slc22a7a | -0.070 |
| tmem243b | 0.169 | cyb5a | -0.069 |
| sptlc2a | 0.168 | pvalb1 | -0.069 |
| glb1l | 0.168 | marcksl1a | -0.068 |
| kita | 0.167 | tmem130 | -0.066 |
| bhlhe40 | 0.167 | gch2 | -0.065 |
| oca2 | 0.166 | elavl4 | -0.065 |
| tspan36 | 0.165 | si:ch73-1a9.3 | -0.065 |
| klf6a | 0.165 | paics | -0.065 |
| arhgef33 | 0.164 | sox11a | -0.064 |
| gpr143 | 0.164 | hmgn2 | -0.064 |
| pcdh10a | 0.163 | hmgb1b | -0.064 |
| si:ch211-195b13.1 | 0.162 | prdx5 | -0.063 |
| rab38 | 0.162 | krt4 | -0.063 |
| slc24a5 | 0.161 | sncb | -0.061 |
| ctsla | 0.161 | epb41a | -0.060 |
| gadd45bb | 0.161 | prdx1 | -0.059 |
| msx1b | 0.159 | mdh1aa | -0.059 |