si:ch211-286b5.5
ZFIN 
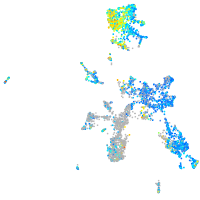
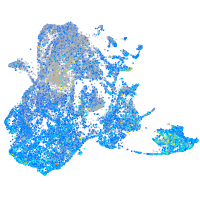
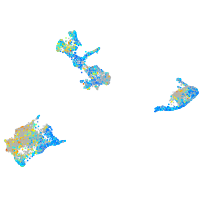
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch1073-429i10.3.1 | 0.571 | ppiaa | -0.558 |
| ptmaa | 0.564 | NC-002333.4 | -0.547 |
| h3f3a | 0.558 | h1m | -0.524 |
| calm1a | 0.547 | chaf1a | -0.517 |
| gapdhs | 0.539 | syncrip | -0.501 |
| wu:fb55g09 | 0.532 | sde2 | -0.481 |
| calm1b | 0.526 | rps18 | -0.478 |
| ccni | 0.525 | ncl | -0.476 |
| smdt1b | 0.518 | ddx5 | -0.476 |
| dap1b | 0.518 | ccna2 | -0.467 |
| ppiab | 0.517 | nop58 | -0.465 |
| rpl37 | 0.510 | cd2bp2 | -0.464 |
| smdt1a | 0.503 | ppig | -0.463 |
| ttr | 0.502 | ctsla | -0.461 |
| apoa1b | 0.500 | g3bp1 | -0.456 |
| tagapa | 0.500 | hnrnpub | -0.454 |
| atp8a2 | 0.499 | sf3b2 | -0.448 |
| tox | 0.493 | stmn1a | -0.448 |
| pip4p1b | 0.480 | ddx4 | -0.444 |
| pkma | 0.478 | mphosph10 | -0.439 |
| tpi1b | 0.478 | zmat2 | -0.438 |
| cst14b.1 | 0.477 | kri1 | -0.437 |
| mir375-2 | 0.475 | hnrnpc | -0.435 |
| myl10 | 0.474 | nasp | -0.434 |
| ckmb | 0.472 | npm1a | -0.431 |
| celf4 | 0.471 | nop56 | -0.427 |
| si:dkey-208c12.2 | 0.468 | rbm7 | -0.420 |
| rps17 | 0.468 | hnrnpa1b | -0.415 |
| acta1a | 0.466 | srsf3b | -0.415 |
| tgfbi | 0.465 | morf4l1 | -0.410 |
| rtn1a | 0.464 | pttg1 | -0.409 |
| faua | 0.463 | srrt | -0.408 |
| im:7152348 | 0.462 | cdk11b | -0.404 |
| vdac2 | 0.460 | stm | -0.398 |
| apoa2 | 0.459 | dnajc1 | -0.397 |