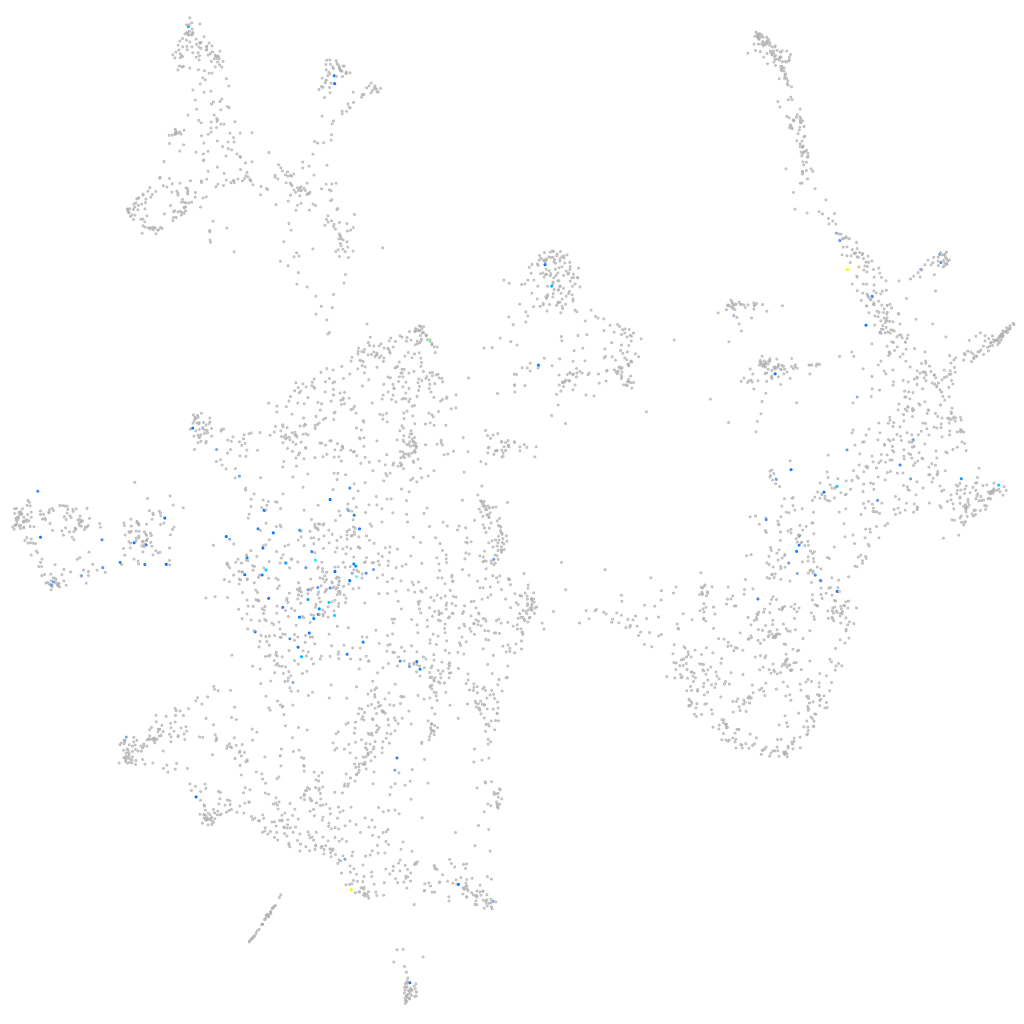
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mad2l1 | 0.181 | zgc:158463 | -0.062 |
| LOC799983 | 0.178 | CR383676.1 | -0.058 |
| LOC100148591 | 0.174 | mt-co2 | -0.056 |
| cks1b | 0.168 | COX3 | -0.052 |
| epha8 | 0.164 | gabarapa | -0.051 |
| BX908744.1 | 0.164 | tob1b | -0.049 |
| znf1132 | 0.164 | socs3a | -0.049 |
| mki67 | 0.161 | ical1 | -0.048 |
| tnni2a.3 | 0.160 | LOC110439372 | -0.045 |
| LOC100330864 | 0.157 | ckma | -0.044 |
| rrm1 | 0.153 | tmem59 | -0.044 |
| ndc80 | 0.150 | abcc5 | -0.043 |
| fhl1a | 0.150 | dedd1 | -0.042 |
| si:dkey-261m9.17 | 0.150 | arglu1a | -0.042 |
| spc25 | 0.150 | cebpd | -0.042 |
| polq | 0.150 | socs3b | -0.041 |
| ccna2 | 0.149 | pvalb1 | -0.040 |
| si:dkey-6i22.5 | 0.147 | atp2b1a | -0.040 |
| cdk1 | 0.146 | gapdhs | -0.040 |
| lbr | 0.145 | gsto2 | -0.040 |
| si:dkey-108k21.14 | 0.145 | gpam | -0.040 |
| tpx2 | 0.145 | pvalb2 | -0.039 |
| CR927050.1 | 0.145 | tspan17 | -0.039 |
| zgc:165555.3 | 0.142 | eya4 | -0.039 |
| ube2t | 0.141 | mid1ip1b | -0.039 |
| zgc:110216 | 0.141 | vamp2 | -0.039 |
| smc2 | 0.141 | eno1a | -0.038 |
| haus1 | 0.140 | h1f0 | -0.038 |
| stmn1a | 0.140 | ctsf | -0.038 |
| XLOC-025418 | 0.137 | htatip2 | -0.038 |
| LOC108182870 | 0.137 | ip6k2a | -0.038 |
| top2a | 0.136 | rab11al | -0.038 |
| fbxo5 | 0.136 | kdm6bb | -0.038 |
| dut | 0.136 | myh14 | -0.037 |
| btr16.1 | 0.135 | RAMP1 | -0.037 |