si:ch211-262h13.5
ZFIN 
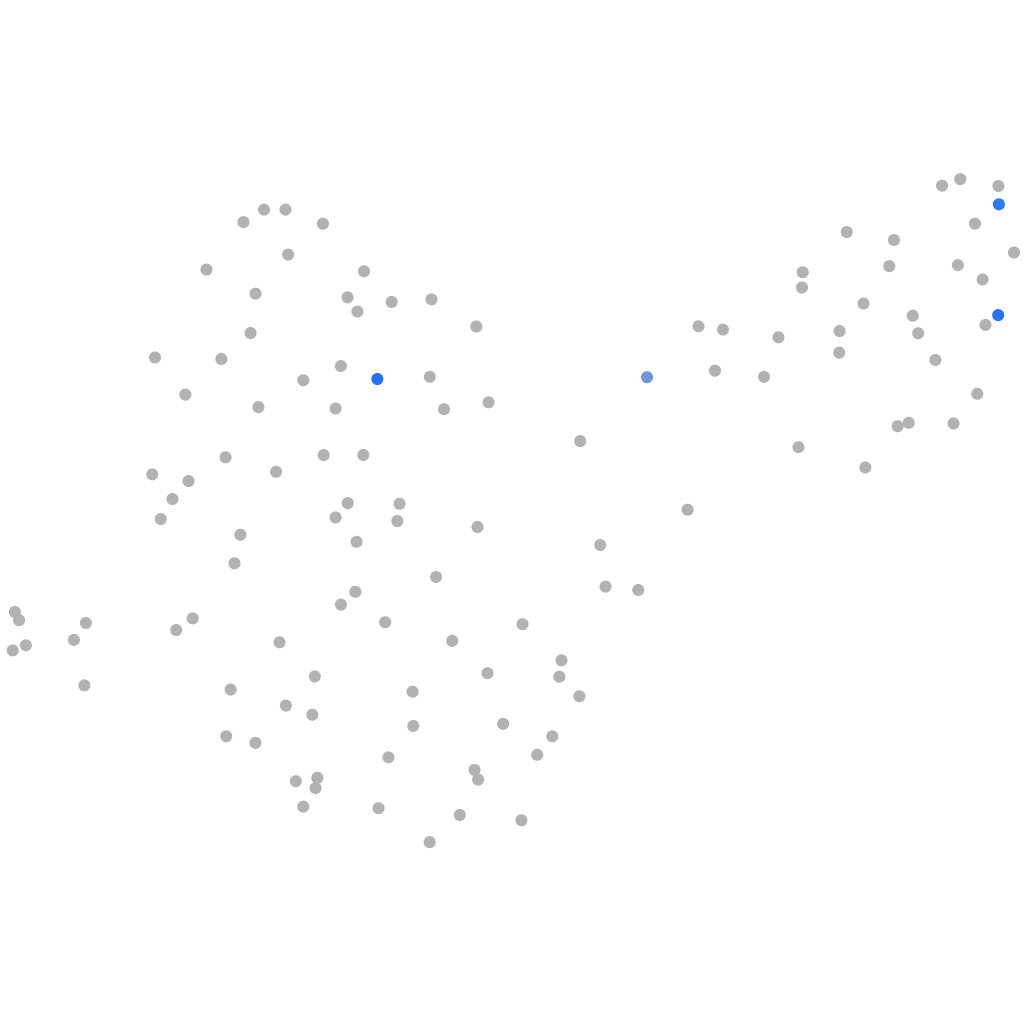
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| swsap1 | 0.919 | ncl | -0.226 |
| elob | 0.786 | hmgb2a | -0.190 |
| glsl | 0.764 | srsf1a | -0.186 |
| nefmb | 0.730 | cbx5 | -0.178 |
| fbxl15 | 0.675 | ddx61 | -0.174 |
| XLOC-011126 | 0.669 | acin1b | -0.170 |
| si:dkey-266m15.6 | 0.639 | banf1 | -0.169 |
| six6b | 0.639 | akap12b | -0.162 |
| LOC101882492 | 0.639 | hmga1a | -0.160 |
| gipr | 0.632 | gtf2f1 | -0.149 |
| igf2a | 0.632 | ctcf | -0.149 |
| myocd | 0.632 | znf346 | -0.147 |
| si:ch211-141o9.10 | 0.632 | si:dkeyp-69c1.6 | -0.147 |
| CR847530.1 | 0.623 | zc3h15 | -0.146 |
| FP067440.1 | 0.610 | nop56 | -0.146 |
| amy2a | 0.606 | smarca4a | -0.145 |
| zdhhc15a | 0.605 | appbp2 | -0.144 |
| coro1a | 0.600 | ywhae1 | -0.143 |
| trim25l | 0.594 | NC-002333.4 | -0.142 |
| sqlea | 0.582 | CT030188.1 | -0.141 |
| lad1 | 0.562 | yy1a | -0.140 |
| adamtsl5 | 0.558 | marcksl1b | -0.139 |
| ak7b | 0.558 | larp7 | -0.139 |
| arhgef10lb | 0.558 | marcksb | -0.137 |
| btr25 | 0.558 | apoc1 | -0.137 |
| BX323800.1 | 0.558 | cherp | -0.136 |
| BX640398.1 | 0.558 | cbx3a | -0.136 |
| BX649398.2 | 0.558 | zgc:77262 | -0.135 |
| BX927253.1 | 0.558 | serbp1b | -0.135 |
| CABZ01011209.1 | 0.558 | ppp1r7 | -0.135 |
| CABZ01012885.1 | 0.558 | zgc:110425 | -0.134 |
| CABZ01053221.1 | 0.558 | cnot6a | -0.133 |
| CABZ01074085.1 | 0.558 | baz1b | -0.132 |
| cacna2d2b | 0.558 | s100a1 | -0.131 |
| calcrlb | 0.558 | nob1 | -0.130 |