si:ch211-207d6.2
ZFIN 
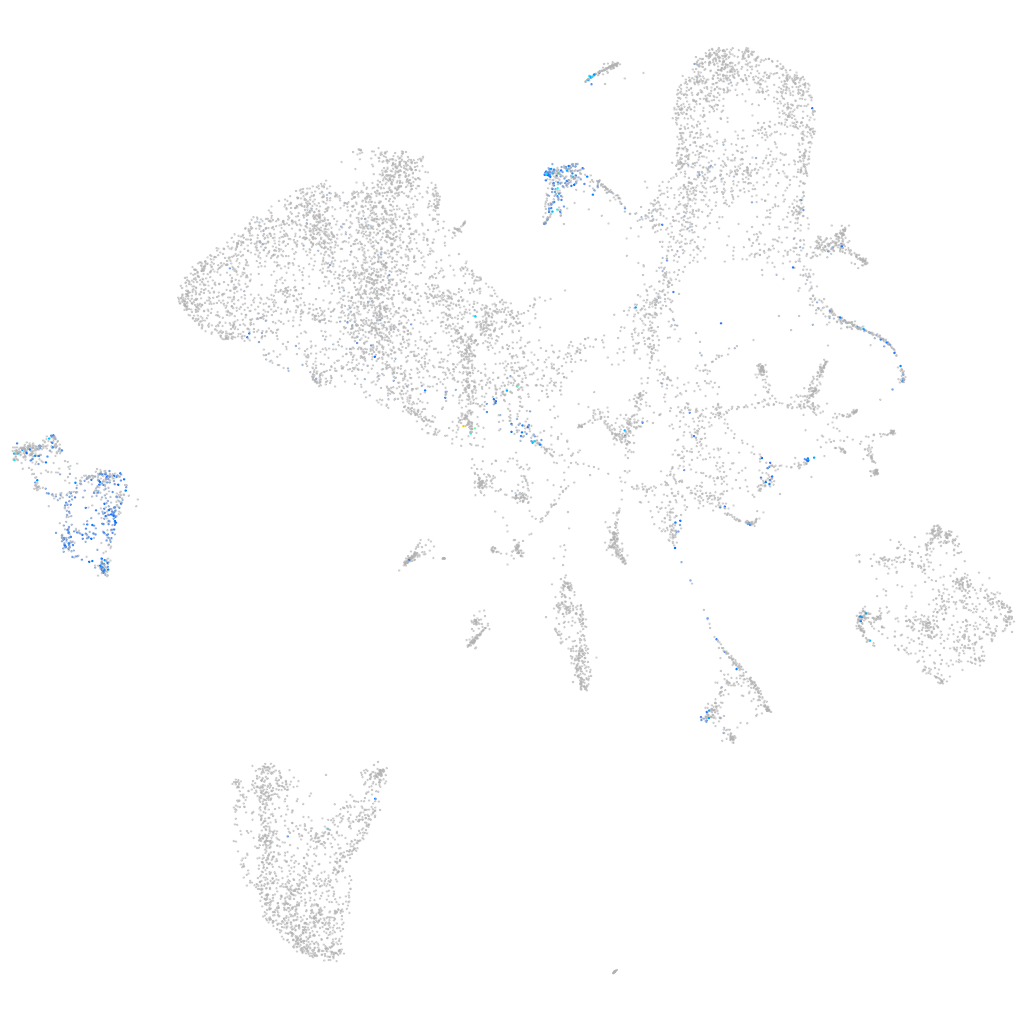
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| s100w | 0.374 | gapdh | -0.174 |
| si:dkey-19b23.12 | 0.373 | gamt | -0.165 |
| zgc:165423 | 0.350 | gatm | -0.163 |
| muc5.3 | 0.336 | apoc2 | -0.149 |
| s100a10b | 0.331 | fbp1b | -0.147 |
| myof | 0.331 | apoa4b.1 | -0.145 |
| cldnh | 0.329 | afp4 | -0.141 |
| anxa3a | 0.328 | gpx4a | -0.139 |
| si:ch211-105c13.3 | 0.326 | bhmt | -0.137 |
| icn | 0.325 | pnp4b | -0.133 |
| CABZ01039859.1 | 0.320 | mat1a | -0.131 |
| gstp2 | 0.319 | apobb.1 | -0.131 |
| ahnak | 0.313 | dhrs9 | -0.129 |
| zgc:153284 | 0.311 | glud1b | -0.128 |
| CABZ01068499.1 | 0.310 | apoc1 | -0.128 |
| zgc:92380 | 0.310 | sult2st2 | -0.127 |
| tppp3 | 0.308 | apoa1b | -0.125 |
| capgb | 0.308 | agxtb | -0.124 |
| ponzr5 | 0.307 | abat | -0.124 |
| cldnb | 0.307 | scp2a | -0.123 |
| cfl1l | 0.307 | ahcy | -0.122 |
| scel | 0.301 | gcshb | -0.122 |
| krt91 | 0.296 | cox7a1 | -0.121 |
| si:dkey-248g15.3 | 0.294 | rdh1 | -0.121 |
| krt1-19d | 0.293 | aqp12 | -0.119 |
| s100v2 | 0.291 | tdo2a | -0.117 |
| capn2b | 0.290 | pklr | -0.116 |
| aqp3a | 0.286 | srd5a2a | -0.115 |
| ftr83 | 0.285 | suclg1 | -0.115 |
| agr2 | 0.283 | msrb2 | -0.115 |
| epcam | 0.282 | pcbd1 | -0.115 |
| anxa2a | 0.280 | pla2g12b | -0.114 |
| XLOC-027129 | 0.278 | etnppl | -0.113 |
| tprg1 | 0.277 | nupr1b | -0.113 |
| capns1a | 0.277 | si:dkey-16p21.8 | -0.113 |