si:ch211-203b20.7 [Source:ZFIN;Acc:ZDB-GENE-060503-302]
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural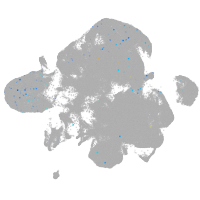 |
spinal cord / glia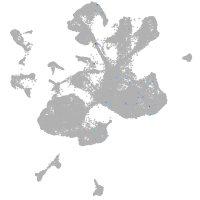 |
eye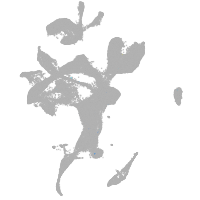 |
taste / olfactory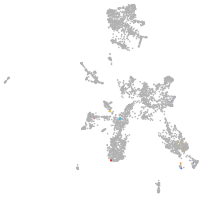 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.047 | gapdhs | -0.020 |
| lig1 | 0.042 | hbae3 | -0.020 |
| BX927258.1 | 0.041 | hbbe1.3 | -0.020 |
| nr6a1a | 0.041 | pvalb1 | -0.020 |
| hnrnpa1b | 0.040 | pvalb2 | -0.020 |
| ppig | 0.040 | cyt1 | -0.019 |
| sp5l | 0.040 | hbae1.1 | -0.019 |
| zgc:110425 | 0.040 | krt4 | -0.019 |
| nop56 | 0.039 | calm1a | -0.019 |
| si:ch211-152c2.3 | 0.039 | hbbe1.1 | -0.018 |
| tpx2 | 0.039 | actc1b | -0.017 |
| dkc1 | 0.038 | atp5if1b | -0.017 |
| mki67 | 0.038 | cap1 | -0.017 |
| nop58 | 0.038 | ccni | -0.017 |
| si:dkey-66i24.9 | 0.038 | ckbb | -0.017 |
| cdca8 | 0.037 | cyt1l | -0.017 |
| cdx4 | 0.037 | ier2b | -0.017 |
| cx43.4 | 0.037 | krtt1c19e | -0.017 |
| fbl | 0.037 | s100a10b | -0.016 |
| ncl | 0.037 | zgc:162730 | -0.016 |
| npm1a | 0.037 | anxa1a | -0.015 |
| srsf1a | 0.037 | atp6v1e1b | -0.015 |
| chaf1a | 0.036 | atp6v1g1 | -0.015 |
| dnmt3bb.2 | 0.036 | cebpd | -0.015 |
| hnrnpub | 0.036 | hbae1.3 | -0.015 |
| nop2 | 0.036 | hbbe2 | -0.015 |
| ppm1g | 0.036 | icn | -0.015 |
| seta | 0.036 | icn2 | -0.015 |
| vrtn | 0.036 | tmsb1 | -0.015 |
| banf1 | 0.035 | tob1b | -0.015 |
| bms1 | 0.035 | tpi1b | -0.015 |
| cdx1a | 0.035 | ahnak | -0.014 |
| ebna1bp2 | 0.035 | atp6v0cb | -0.014 |
| gnl3 | 0.035 | col1a2 | -0.014 |
| rbm4.3 | 0.035 | hbbe1.2 | -0.014 |




