si:ch211-195d17.2
ZFIN 
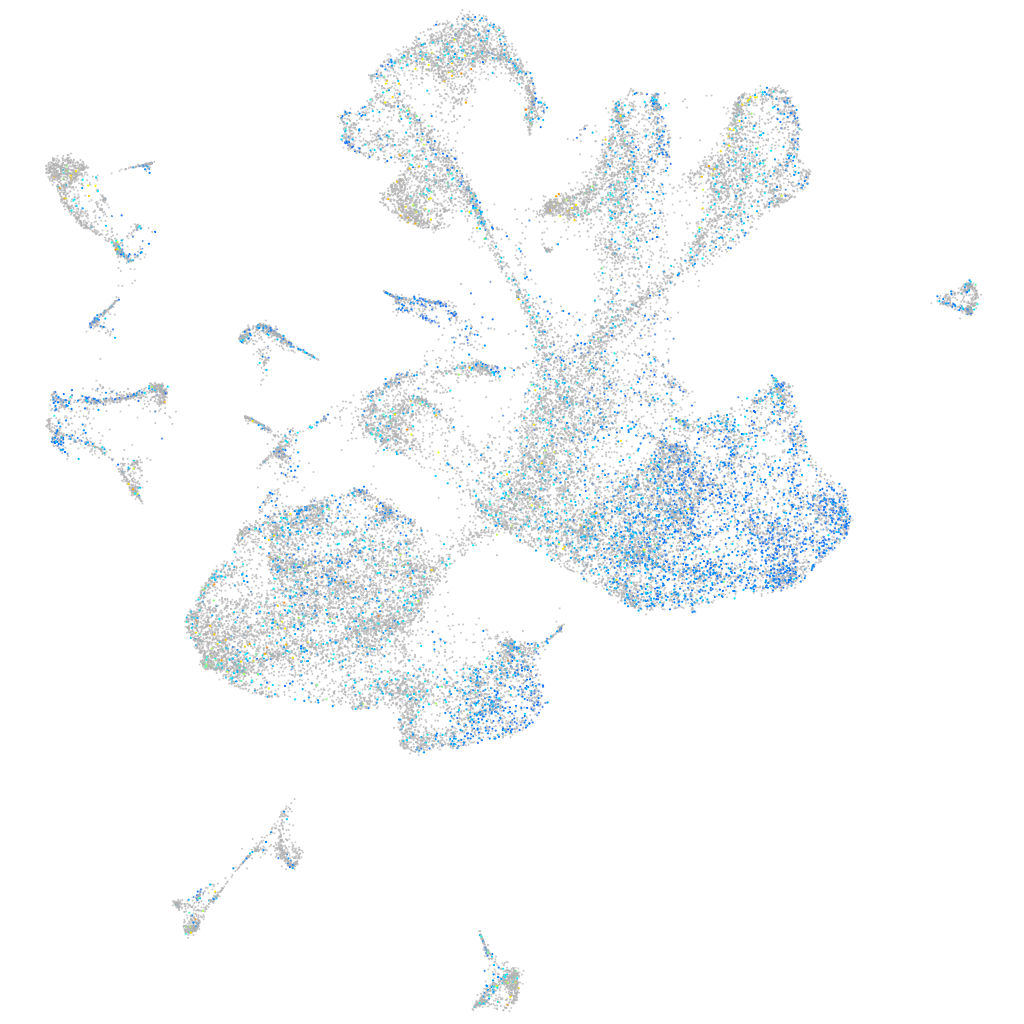
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.119 | COX3 | -0.077 |
| dut | 0.113 | rtn1a | -0.059 |
| selenoh | 0.111 | myt1b | -0.057 |
| nasp | 0.108 | elavl3 | -0.056 |
| anp32b | 0.108 | ckbb | -0.054 |
| fen1 | 0.106 | CU467822.1 | -0.054 |
| stmn1a | 0.103 | gpm6aa | -0.053 |
| chaf1a | 0.103 | tmsb | -0.053 |
| rpa2 | 0.103 | nova2 | -0.051 |
| mcm7 | 0.103 | gpm6ab | -0.050 |
| cbx3a | 0.103 | actc1b | -0.046 |
| zgc:56493 | 0.102 | hbae3 | -0.046 |
| rpa3 | 0.101 | pvalb1 | -0.046 |
| hmgb2a | 0.101 | mt-co2 | -0.046 |
| cnbpa | 0.101 | stmn1b | -0.046 |
| ran | 0.100 | pvalb2 | -0.046 |
| rrm1 | 0.100 | srrm4 | -0.045 |
| cks1b | 0.100 | CU634008.1 | -0.045 |
| tuba8l4 | 0.097 | slc1a2b | -0.045 |
| rnaseh2a | 0.096 | marcksl1b | -0.044 |
| zgc:110540 | 0.096 | hbbe1.3 | -0.044 |
| nutf2l | 0.096 | zgc:158463 | -0.041 |
| dek | 0.094 | mt-co1 | -0.040 |
| npm1a | 0.094 | hbbe1.1 | -0.040 |
| lig1 | 0.094 | CR848047.1 | -0.039 |
| slbp | 0.094 | scrt2 | -0.039 |
| LOC100330864 | 0.092 | mdkb | -0.038 |
| unga | 0.092 | ebf3a | -0.037 |
| dhfr | 0.092 | scrt1a | -0.037 |
| rrm2 | 0.092 | ptn | -0.037 |
| ranbp1 | 0.092 | myt1a | -0.036 |
| h2afvb | 0.091 | elavl4 | -0.036 |
| si:ch73-281n10.2 | 0.091 | nfia | -0.036 |
| banf1 | 0.091 | ube2d4 | -0.036 |
| ccna2 | 0.091 | tmsb4x | -0.036 |