si:ch211-195b15.8
ZFIN 
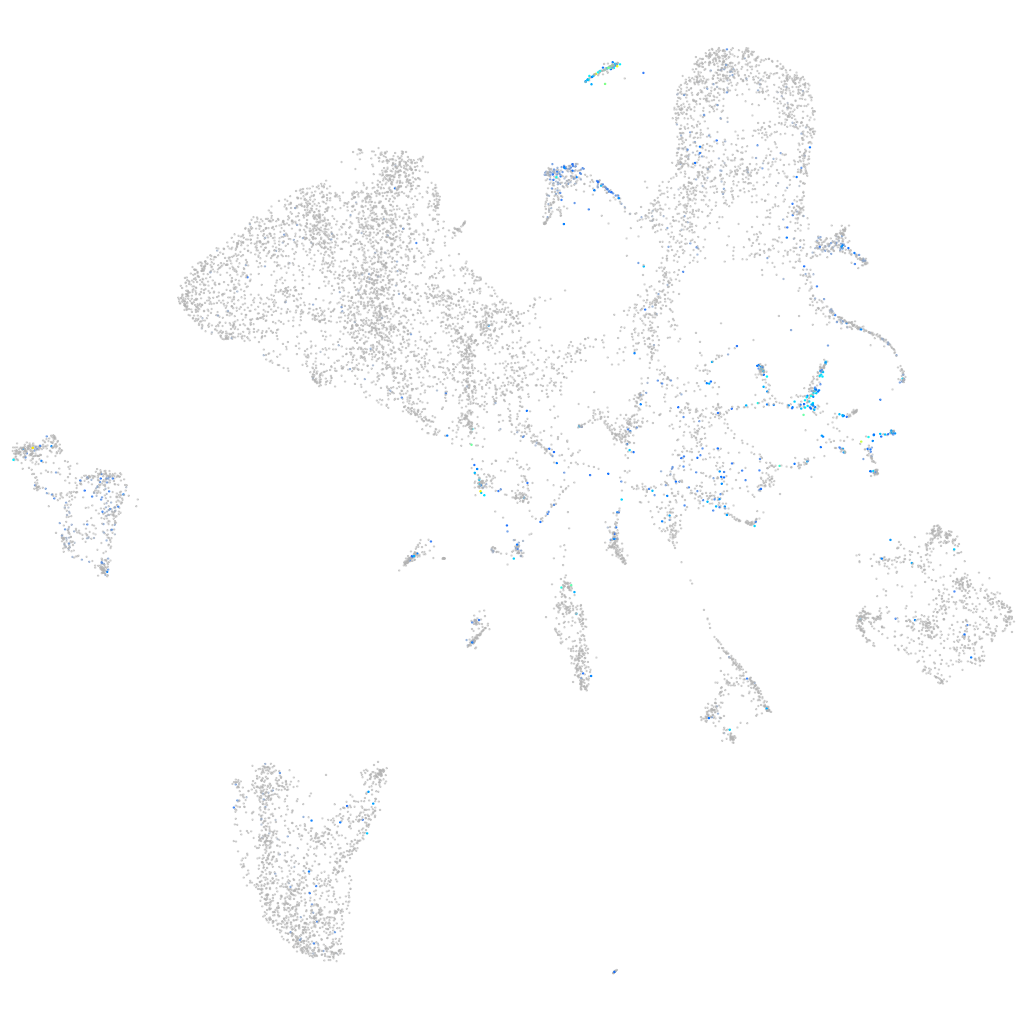
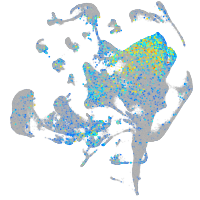
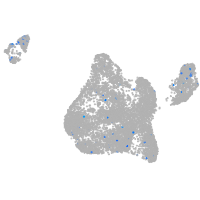
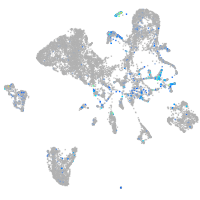
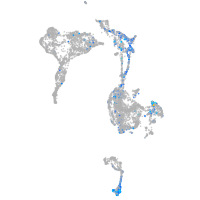
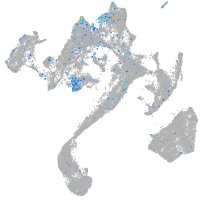
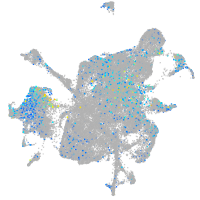
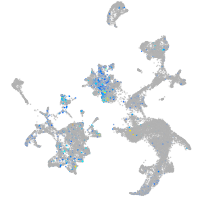
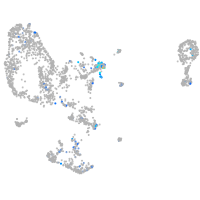
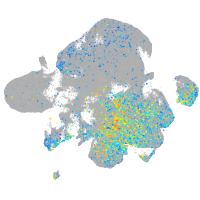
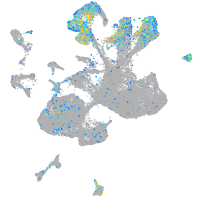
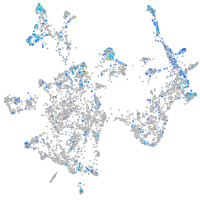
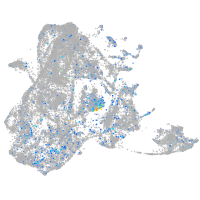
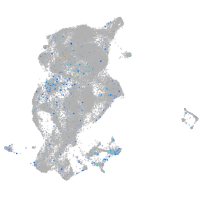
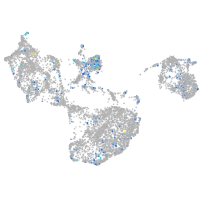
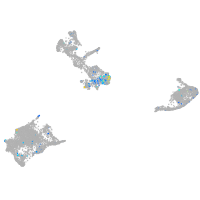
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.360 | gapdh | -0.170 |
| egr4 | 0.337 | gamt | -0.170 |
| c2cd4a | 0.335 | ahcy | -0.168 |
| zgc:101731 | 0.320 | gatm | -0.152 |
| mir7a-1 | 0.296 | mat1a | -0.145 |
| pcsk1nl | 0.292 | bhmt | -0.143 |
| neurod1 | 0.284 | apoa1b | -0.143 |
| calm1b | 0.281 | fbp1b | -0.141 |
| hepacam2 | 0.275 | apoa4b.1 | -0.141 |
| sprn2 | 0.275 | apoc2 | -0.140 |
| elovl1a | 0.267 | agxtb | -0.137 |
| adcyap1a | 0.267 | gstt1a | -0.136 |
| gapdhs | 0.266 | scp2a | -0.134 |
| fev | 0.265 | aldh6a1 | -0.133 |
| insm1b | 0.263 | aldh7a1 | -0.132 |
| syt1a | 0.261 | afp4 | -0.130 |
| insm1a | 0.258 | agxta | -0.130 |
| mir375-2 | 0.257 | aqp12 | -0.129 |
| rgs16 | 0.256 | si:dkey-16p21.8 | -0.128 |
| syt11a | 0.255 | apoc1 | -0.128 |
| id4 | 0.253 | mgst1.2 | -0.128 |
| tox | 0.252 | ttc36 | -0.127 |
| nmbb | 0.249 | rbp2b | -0.126 |
| chga | 0.249 | glud1b | -0.126 |
| vamp2 | 0.246 | tfa | -0.126 |
| pcsk1 | 0.245 | pnp4b | -0.125 |
| si:dkey-153k10.9 | 0.245 | cx32.3 | -0.125 |
| mapk15 | 0.243 | serpina1 | -0.125 |
| ier2b | 0.241 | ftcd | -0.124 |
| lrrc39 | 0.239 | fabp10a | -0.124 |
| anxa4 | 0.238 | hpda | -0.123 |
| vat1 | 0.237 | serpina1l | -0.123 |
| hsbp1a | 0.236 | uox | -0.122 |
| scgn | 0.234 | apoa2 | -0.121 |
| nkx2.2a | 0.234 | fabp3 | -0.121 |