si:ch211-191c10.1
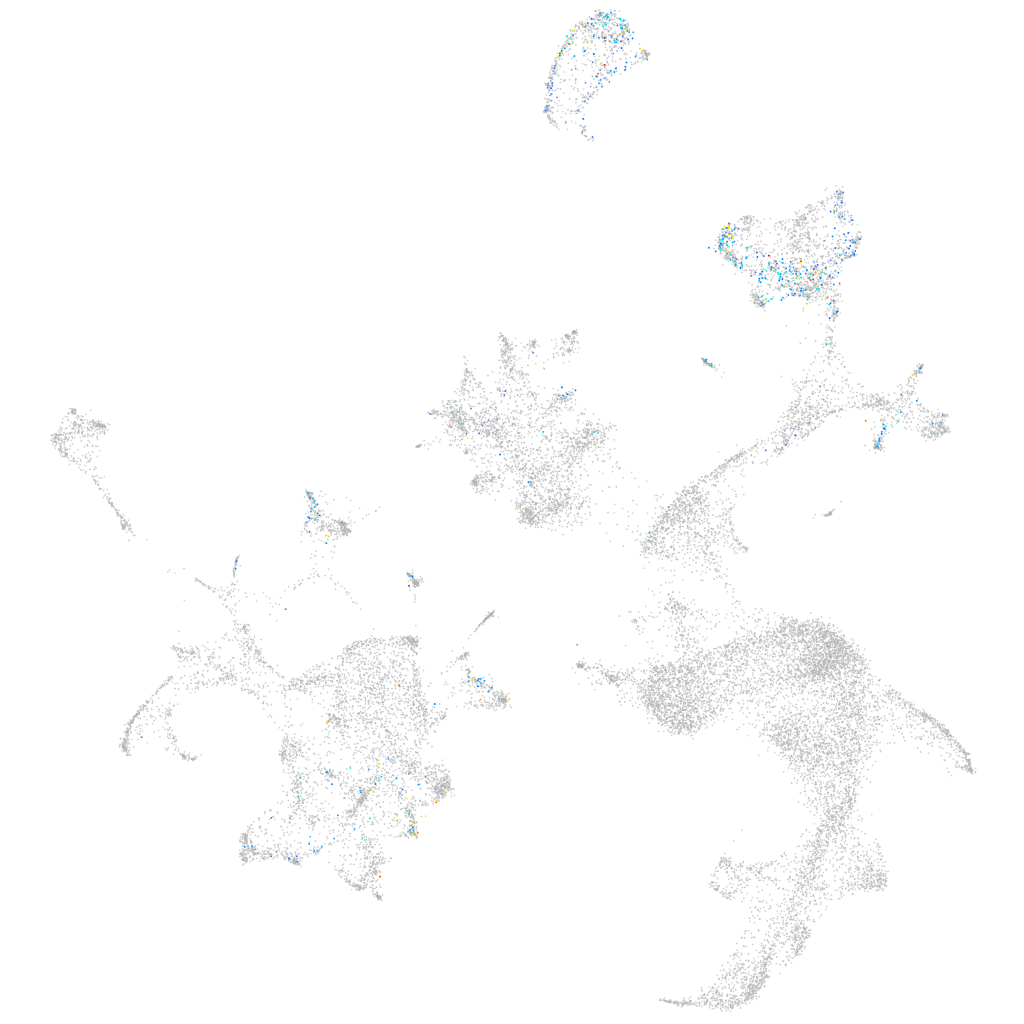
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 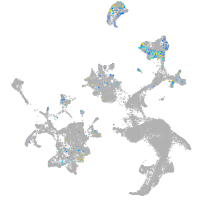 |
pronephros  |
neural |
spinal cord / glia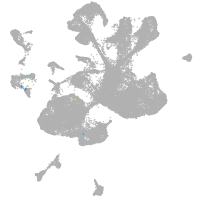 |
eye |
taste / olfactory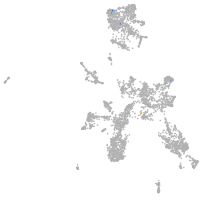 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous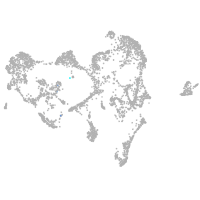 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fcer1gl | 0.224 | hbae1.1 | -0.138 |
| cmklr1 | 0.215 | hbae3 | -0.133 |
| laptm5 | 0.215 | hbbe1.3 | -0.130 |
| havcr1 | 0.211 | hbae1.3 | -0.129 |
| si:zfos-741a10.3 | 0.206 | hbbe2 | -0.128 |
| ctss2.1 | 0.205 | hbbe1.1 | -0.123 |
| si:ch211-212k18.7 | 0.205 | cahz | -0.122 |
| mpeg1.1 | 0.204 | fth1a | -0.116 |
| si:ch211-102c2.4 | 0.202 | hbbe1.2 | -0.116 |
| si:ch211-241f5.3 | 0.199 | hemgn | -0.113 |
| cebpb | 0.198 | zgc:56095 | -0.110 |
| itgb2 | 0.198 | alas2 | -0.107 |
| arpc1b | 0.196 | blvrb | -0.104 |
| il1b | 0.194 | si:ch211-250g4.3 | -0.103 |
| cx32.2 | 0.191 | slc4a1a | -0.101 |
| zgc:64051 | 0.189 | nt5c2l1 | -0.100 |
| itgae.1 | 0.188 | epb41b | -0.095 |
| slc7a7 | 0.187 | creg1 | -0.095 |
| lgals3bpb | 0.184 | zgc:163057 | -0.094 |
| dusp2 | 0.183 | si:ch211-207c6.2 | -0.091 |
| ctsc | 0.179 | nmt1b | -0.087 |
| adgrg3 | 0.179 | tspo | -0.085 |
| CU499330.1 | 0.178 | prdx2 | -0.082 |
| ptprc | 0.177 | lmo2 | -0.082 |
| marco | 0.177 | tmod4 | -0.082 |
| si:dkey-5n18.1 | 0.177 | plac8l1 | -0.081 |
| zgc:162730 | 0.177 | uros | -0.077 |
| vsir | 0.177 | hbae5 | -0.076 |
| spi1b | 0.176 | znfl2a | -0.076 |
| ccl34b.1 | 0.176 | sptb | -0.075 |
| tnfb | 0.175 | tfr1a | -0.075 |
| lect2l | 0.172 | hbbe3 | -0.074 |
| nfkbiaa | 0.172 | anp32b | -0.073 |
| coro1a | 0.171 | rfesd | -0.072 |
| pfn1 | 0.171 | hdr | -0.071 |