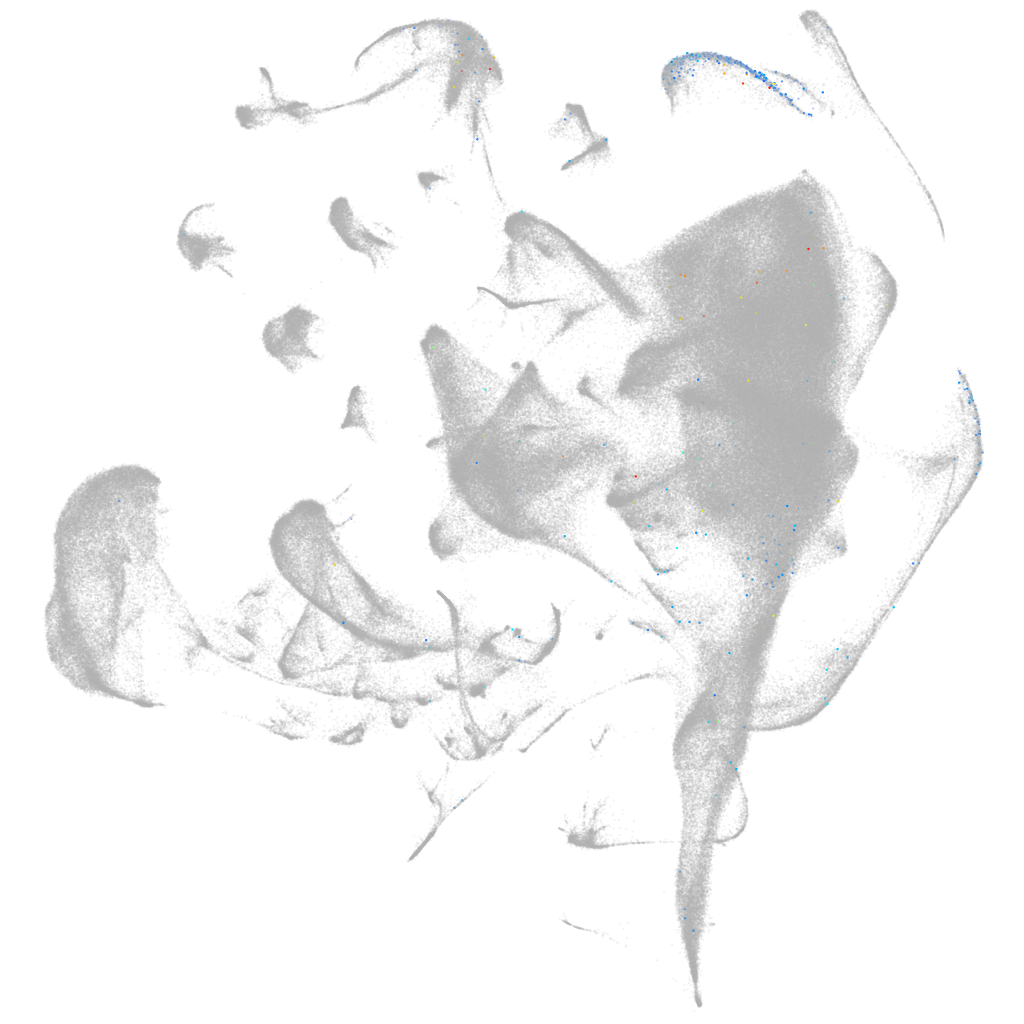si:ch211-158m24.12
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch1073-100f3.2 | 0.216 | hmgb1b | -0.027 |
| rtn2b | 0.212 | si:ch211-288g17.3 | -0.027 |
| tgm2a | 0.209 | ywhaqb | -0.026 |
| cavin4a | 0.203 | marcksl1a | -0.026 |
| myoz1b | 0.202 | h2afva | -0.025 |
| myoz2a | 0.200 | ckbb | -0.024 |
| hhatla | 0.195 | ctsla | -0.024 |
| myhc4 | 0.195 | ddx5 | -0.024 |
| tnnt3a | 0.195 | gnb1b | -0.024 |
| CABZ01078594.1 | 0.194 | gpm6aa | -0.024 |
| casq1a | 0.193 | tmem258 | -0.024 |
| cdnf | 0.192 | ywhabb | -0.024 |
| prr33 | 0.186 | chd4a | -0.023 |
| zgc:158296 | 0.186 | gapdhs | -0.023 |
| mybpc2b | 0.185 | nova2 | -0.023 |
| capn3b | 0.183 | pfn2 | -0.023 |
| myoz1a | 0.183 | tuba1c | -0.023 |
| xirp2b | 0.183 | zgc:153867 | -0.023 |
| ryr1b | 0.181 | anp32a | -0.022 |
| cavin4b | 0.180 | hdac1 | -0.022 |
| pgam2 | 0.179 | mdkb | -0.022 |
| myom2a | 0.178 | rnasekb | -0.022 |
| mybphb | 0.177 | si:dkey-56m19.5 | -0.022 |
| smyd1a | 0.177 | acin1a | -0.021 |
| cacna1sb | 0.176 | atrx | -0.021 |
| cav3 | 0.176 | marcksb | -0.021 |
| ldb3b | 0.175 | srsf3b | -0.021 |
| cx39.9 | 0.174 | zc4h2 | -0.021 |
| myhz2 | 0.174 | cbx5 | -0.020 |
| casq1b | 0.173 | gpm6ab | -0.020 |
| dhrs7cb | 0.173 | hmgb3a | -0.020 |
| si:dkey-211f22.5 | 0.173 | hnrnpa0a | -0.020 |
| myl1 | 0.172 | nucks1a | -0.020 |
| myom1a | 0.172 | pdia3 | -0.020 |
| tmem182a | 0.172 | si:dkey-276j7.1 | -0.020 |