si:ch211-130m23.5
ZFIN 
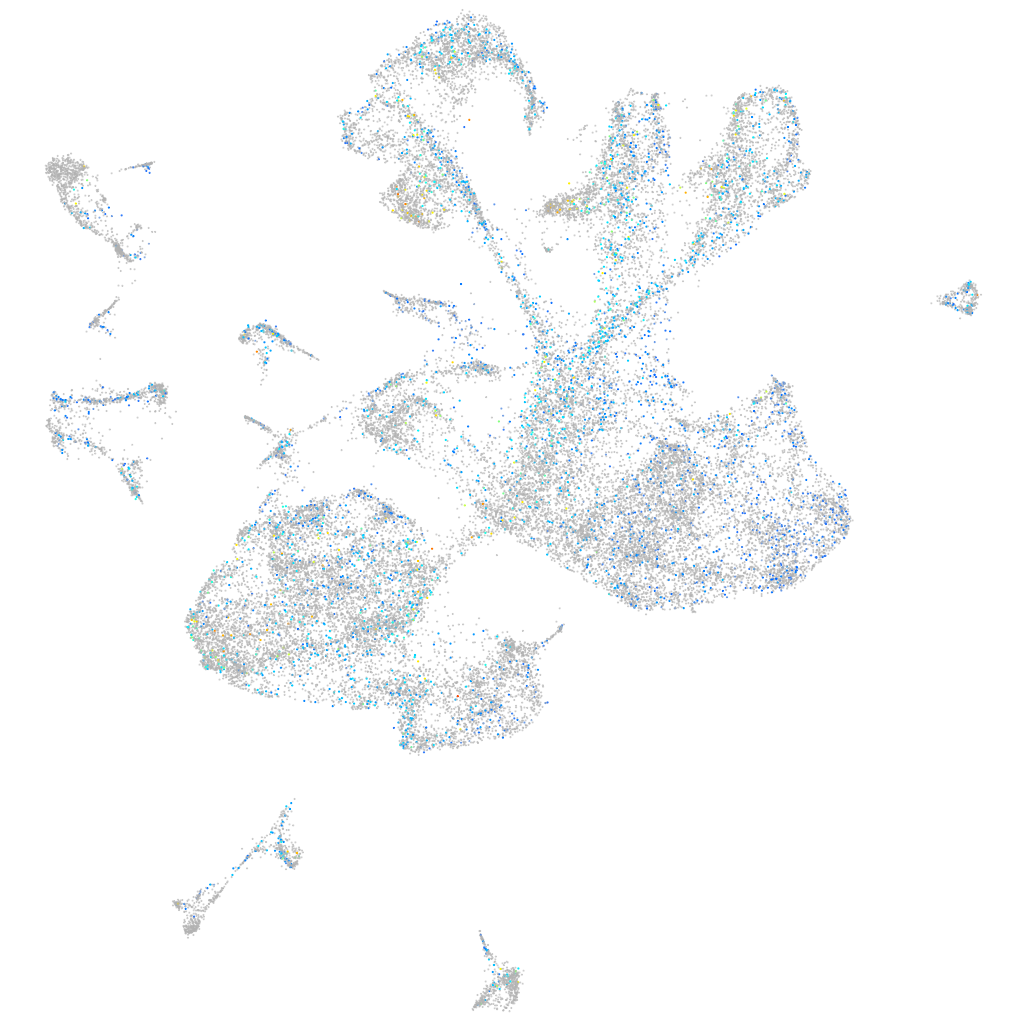
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nhlh2 | 0.089 | mki67 | -0.048 |
| elavl3 | 0.086 | dut | -0.045 |
| myt1b | 0.084 | lbr | -0.045 |
| ebf2 | 0.079 | pcna | -0.043 |
| myt1a | 0.078 | dlgap5 | -0.043 |
| dlb | 0.077 | cdc20 | -0.042 |
| rtn1a | 0.076 | dek | -0.042 |
| scrt2 | 0.076 | ccnb1 | -0.042 |
| ebf3a | 0.075 | mad2l1 | -0.042 |
| insm1b | 0.075 | aspm | -0.041 |
| nova2 | 0.074 | tuba8l | -0.041 |
| jpt1b | 0.072 | zwi | -0.040 |
| sox11b | 0.070 | tpx2 | -0.040 |
| tubb5 | 0.069 | cks1b | -0.039 |
| hmgb3a | 0.069 | hmgb2a | -0.039 |
| neurod1 | 0.068 | ncapg | -0.039 |
| tmeff1b | 0.068 | stmn1a | -0.039 |
| scrt1a | 0.068 | nusap1 | -0.038 |
| tuba1c | 0.066 | cdk1 | -0.038 |
| si:dkey-56m19.5 | 0.066 | top2a | -0.038 |
| hnrnpa0a | 0.066 | ube2c | -0.038 |
| lima1a | 0.066 | spdl1 | -0.038 |
| insm1a | 0.066 | her2 | -0.038 |
| si:dkey-28b4.7 | 0.065 | ptgr1 | -0.037 |
| srrm4 | 0.064 | aurka | -0.037 |
| stap2a | 0.064 | cldnk | -0.037 |
| fscn1a | 0.063 | ccna2 | -0.037 |
| tmsb | 0.063 | kifc1 | -0.037 |
| elocb | 0.062 | g2e3 | -0.037 |
| atcaya | 0.062 | cenpf | -0.036 |
| sept5a | 0.062 | ccnd1 | -0.036 |
| ywhah | 0.062 | banf1 | -0.036 |
| gabarapb | 0.061 | si:ch211-286o17.1 | -0.036 |
| si:ch211-133n4.4 | 0.060 | aurkb | -0.036 |
| ubc | 0.060 | plk1 | -0.035 |