si:ch211-12h2.8
ZFIN 
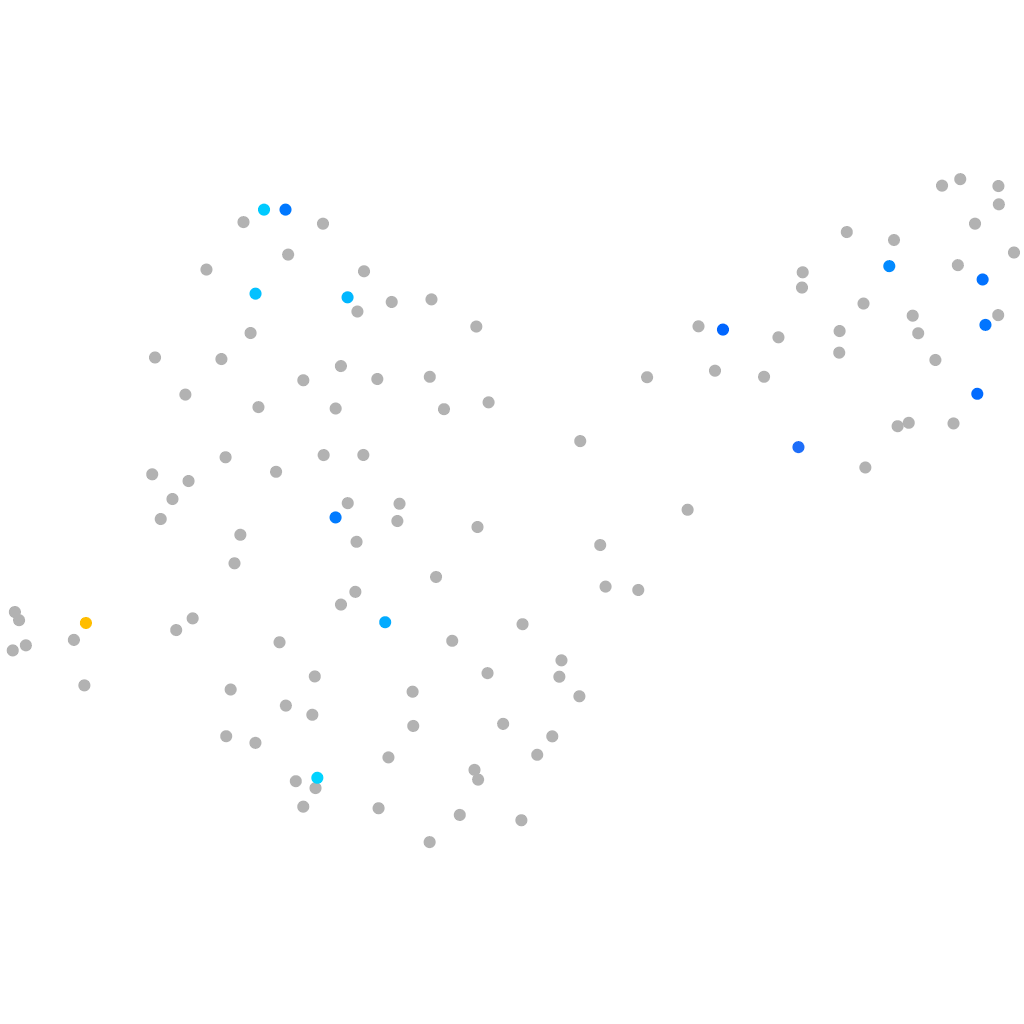
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 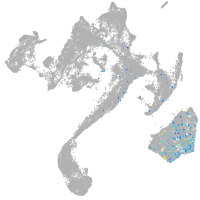 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 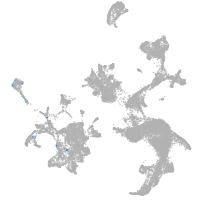 |
pronephros  |
neural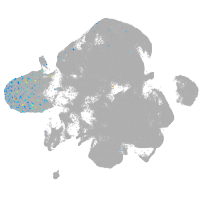 |
spinal cord / glia |
eye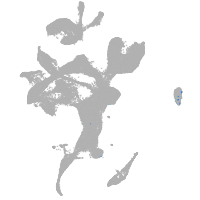 |
taste / olfactory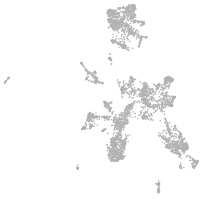 |
otic / lateral line |
epidermis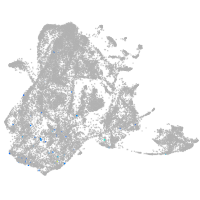 |
periderm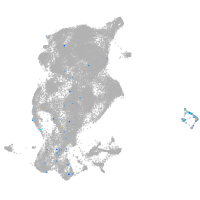 |
fin |
ionocytes / mucous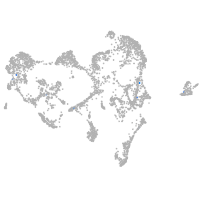 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| VIT | 0.582 | ddit4 | -0.218 |
| smtnl | 0.579 | kmt2bb | -0.199 |
| snapc2 | 0.575 | rpl36 | -0.196 |
| acsl2 | 0.575 | ik | -0.194 |
| dlx4a | 0.575 | mrpl19 | -0.187 |
| he1.2 | 0.575 | naca | -0.187 |
| dlx3b | 0.563 | rps13 | -0.187 |
| XLOC-036447 | 0.562 | srp9 | -0.186 |
| si:dkey-33i11.4 | 0.551 | ankhd1 | -0.185 |
| cntfr | 0.546 | rps27.1 | -0.183 |
| si:dkey-14k9.3 | 0.542 | ywhaz | -0.182 |
| bag6l | 0.540 | rpl28 | -0.181 |
| calml4b | 0.529 | rpl36a | -0.180 |
| acot20 | 0.522 | rpl21 | -0.179 |
| LOC110440123 | 0.522 | eif3s10 | -0.179 |
| si:dkey-148f10.4 | 0.519 | mrpl2 | -0.177 |
| ezh1 | 0.511 | wrb | -0.177 |
| itgb6 | 0.502 | tyw1 | -0.176 |
| tuft1b | 0.498 | gigyf2 | -0.175 |
| glcci1a | 0.495 | pdhb | -0.175 |
| znf1117 | 0.491 | mt-co1 | -0.174 |
| peli1b | 0.480 | eif4e2rs1 | -0.173 |
| CU326366.2 | 0.467 | nipblb | -0.171 |
| znf1161 | 0.467 | rps20 | -0.171 |
| pdxp | 0.460 | si:ch211-126c2.4 | -0.171 |
| junbb | 0.458 | poldip3 | -0.170 |
| LOC100148045 | 0.458 | gtpbp1l | -0.166 |
| gata3 | 0.446 | mob1ba | -0.166 |
| jcada | 0.445 | rps26l | -0.165 |
| ern1 | 0.443 | NC-002333.17 | -0.165 |
| ttbk1b | 0.442 | sltm | -0.165 |
| XLOC-022740 | 0.441 | rbb4l | -0.165 |
| plekho1a | 0.439 | dnajc7 | -0.164 |
| CABZ01075242.1 | 0.437 | dolpp1 | -0.164 |
| si:ch73-213k20.5 | 0.435 | pwp1 | -0.164 |