si:ch211-125m10.6
ZFIN 
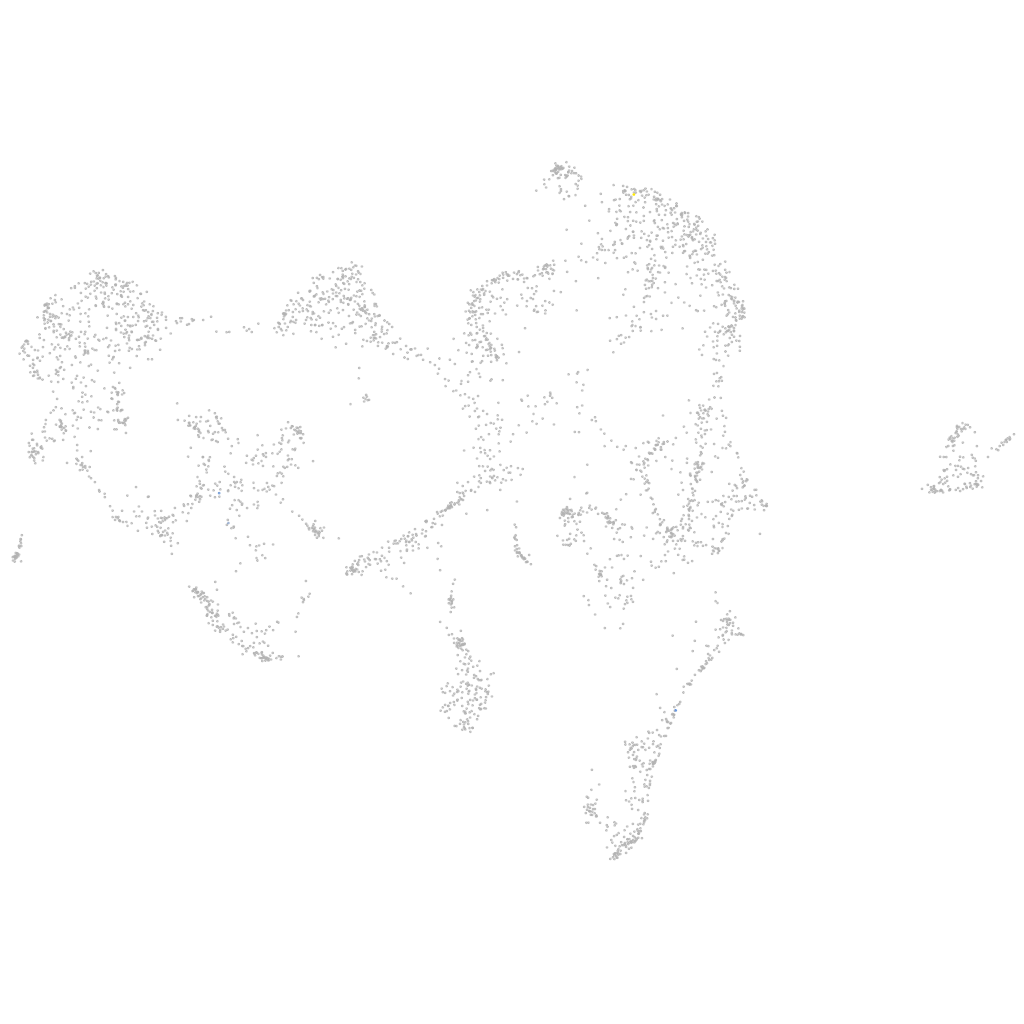
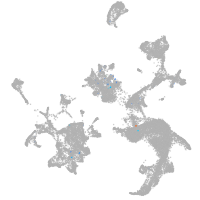
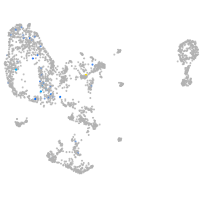
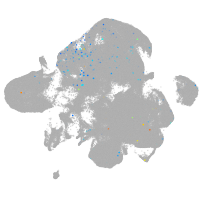
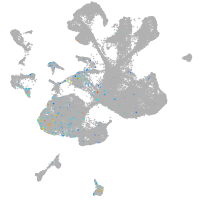
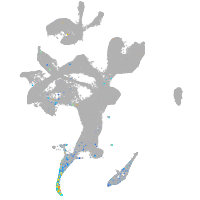
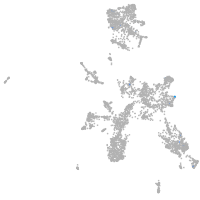
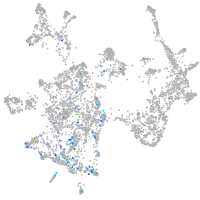
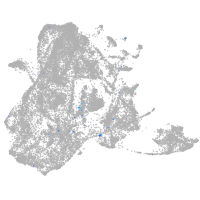
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc6a19a.1 | 0.969 | rpl23a | -0.060 |
| CU466278.1 | 0.851 | rps17 | -0.054 |
| cfh | 0.720 | rpl3 | -0.046 |
| lenep | 0.629 | mt-atp6 | -0.046 |
| slc2a2 | 0.584 | rplp2l | -0.042 |
| dnajc16 | 0.576 | atp5f1e | -0.039 |
| cobl | 0.557 | COX5B | -0.038 |
| LOC110440168 | 0.550 | cox8a | -0.038 |
| hoxa13b | 0.506 | rps18 | -0.037 |
| adat1 | 0.445 | rpl6 | -0.037 |
| LOC103911430 | 0.432 | zgc:56493 | -0.036 |
| LO018166.1 | 0.425 | mt-nd1 | -0.036 |
| dennd4b | 0.348 | cox6a1 | -0.036 |
| smyd1a | 0.335 | atp5pf | -0.035 |
| ASTE1 | 0.332 | ndufa4l | -0.034 |
| b3gnt2a | 0.317 | rpl5b | -0.034 |
| scarb1 | 0.307 | atp5f1b | -0.034 |
| pklr | 0.292 | cox7c | -0.033 |
| CABZ01061524.1 | 0.270 | atp5fa1 | -0.032 |
| rab12 | 0.261 | zgc:193541 | -0.032 |
| myom1a | 0.258 | cox7b | -0.032 |
| mier2 | 0.248 | atp5pd | -0.031 |
| rsf1a | 0.230 | atp5meb | -0.031 |
| agxtb | 0.230 | NC-002333.17 | -0.031 |
| plekhm1 | 0.227 | h3f3c | -0.030 |
| trib2 | 0.222 | pnrc2 | -0.030 |
| serpina1l | 0.222 | hnrnpa0l | -0.030 |
| prmt3 | 0.221 | ndufb8 | -0.029 |
| kank4 | 0.215 | hmgn6 | -0.029 |
| apom | 0.201 | fkbp1aa | -0.029 |
| serac1 | 0.200 | rpl38 | -0.029 |
| rbp2b | 0.194 | atp5f1d | -0.028 |
| pxylp1 | 0.192 | cox5aa | -0.028 |
| rhbdd2 | 0.189 | tma7 | -0.028 |
| ces2 | 0.189 | ndufb9 | -0.028 |