si:ch211-106a19.1
ZFIN 
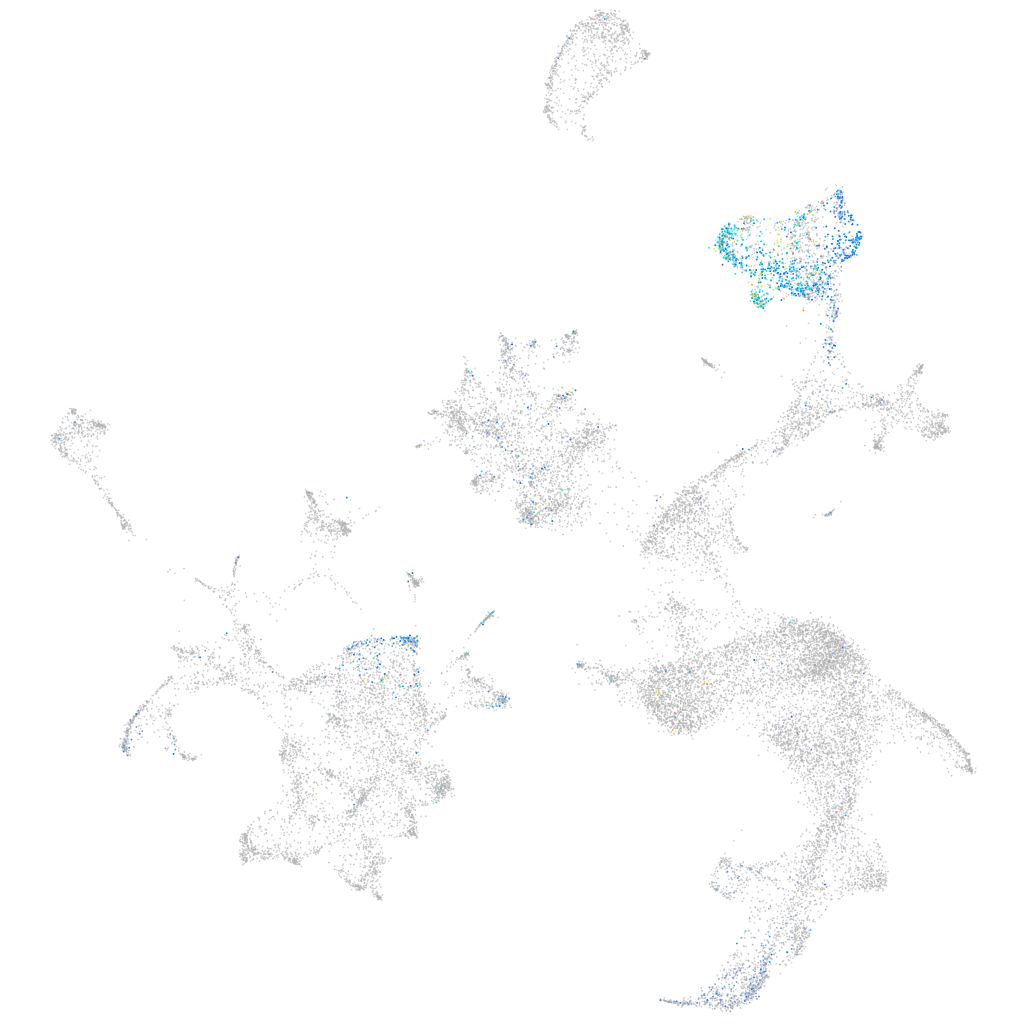
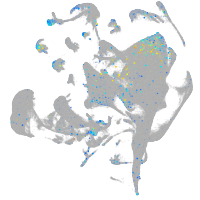
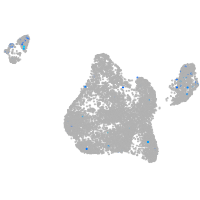
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc7a7 | 0.531 | hbae1.1 | -0.166 |
| havcr1 | 0.528 | hbbe2 | -0.161 |
| mpeg1.1 | 0.517 | hbbe1.3 | -0.159 |
| cmklr1 | 0.515 | hbae3 | -0.158 |
| si:dkey-5n18.1 | 0.485 | hbae1.3 | -0.157 |
| cx32.2 | 0.484 | hbbe1.1 | -0.152 |
| slc3a2a | 0.482 | cahz | -0.149 |
| lgals3bpb | 0.482 | hemgn | -0.140 |
| ctsc | 0.477 | fth1a | -0.139 |
| tspan36 | 0.470 | hbbe1.2 | -0.137 |
| lgals2a | 0.466 | zgc:56095 | -0.136 |
| ctss2.2 | 0.466 | si:ch211-250g4.3 | -0.136 |
| ctsba | 0.464 | alas2 | -0.121 |
| ccl34b.1 | 0.463 | nt5c2l1 | -0.120 |
| mfap4 | 0.458 | creg1 | -0.119 |
| si:zfos-1069f5.1 | 0.453 | slc4a1a | -0.116 |
| mpp1 | 0.451 | si:ch211-207c6.2 | -0.115 |
| si:ch211-212k18.7 | 0.449 | plac8l1 | -0.112 |
| ctsl.1 | 0.448 | tmod4 | -0.108 |
| tnfb | 0.444 | lmo2 | -0.106 |
| tnfsf12 | 0.442 | zgc:163057 | -0.104 |
| si:ch73-158p21.3 | 0.437 | tspo | -0.103 |
| CU499330.1 | 0.428 | blvrb | -0.102 |
| ctsz | 0.413 | epb41b | -0.097 |
| marco | 0.408 | aqp1a.1 | -0.096 |
| lgals9l1 | 0.408 | nmt1b | -0.094 |
| psap | 0.407 | si:dkey-240n22.3 | -0.087 |
| irf8 | 0.406 | si:ch211-222l21.1 | -0.084 |
| scpep1 | 0.405 | histh1l | -0.083 |
| mafbb | 0.404 | hbae5 | -0.083 |
| itgae.1 | 0.403 | add2 | -0.077 |
| ccl35.1 | 0.398 | si:ch211-227m13.1 | -0.077 |
| slc35f6 | 0.396 | sptb | -0.076 |
| rnaset2l | 0.395 | ucp3 | -0.076 |
| cndp2 | 0.394 | uros | -0.073 |