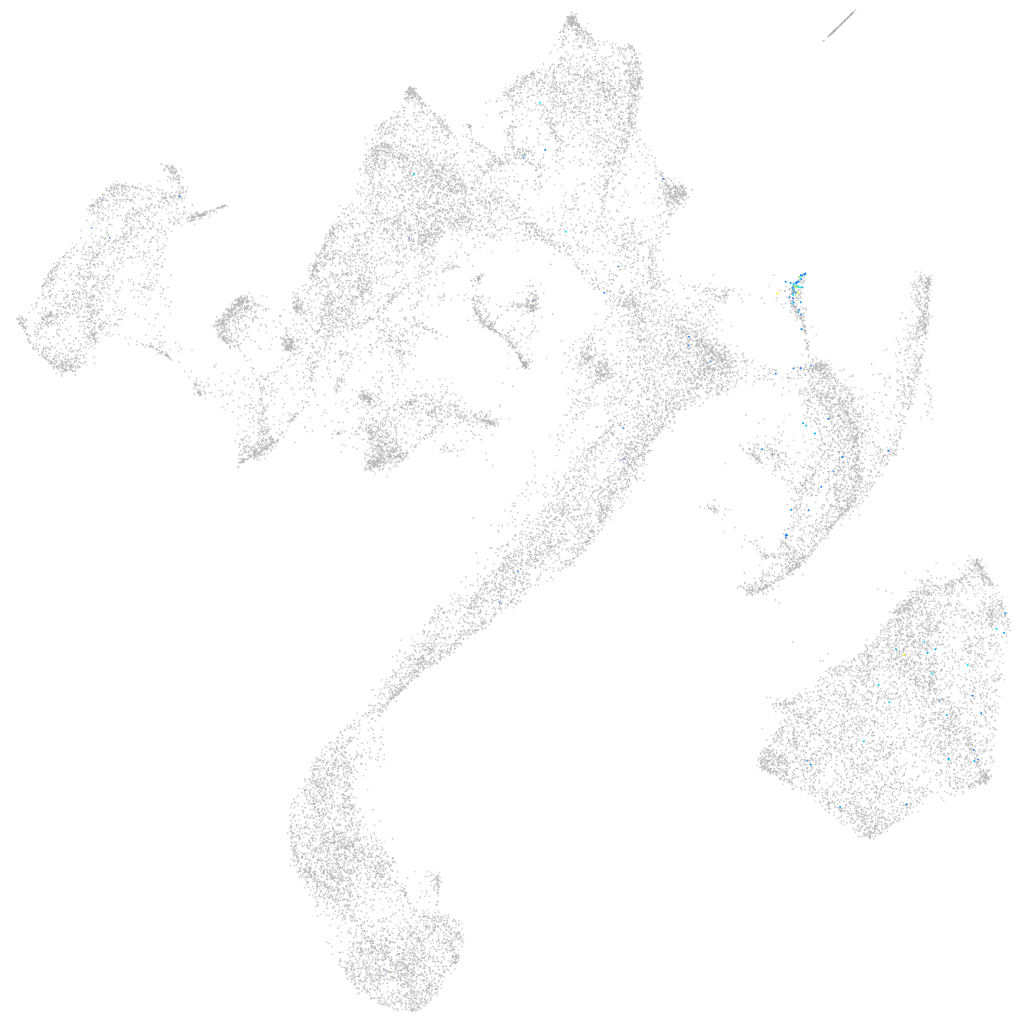
si:cabz01036006.1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hoxa13a | 0.220 | actc1b | -0.040 |
| hoxb13a | 0.190 | ttn.2 | -0.038 |
| hoxc13a | 0.169 | ttn.1 | -0.038 |
| hoxc13b | 0.168 | fabp3 | -0.037 |
| si:dkeyp-110a12.4 | 0.164 | bhmt | -0.034 |
| wnt10a | 0.163 | ak1 | -0.034 |
| hoxa13b | 0.152 | pabpc4 | -0.033 |
| LOC110439452 | 0.139 | aldoab | -0.033 |
| XLOC-004335 | 0.123 | tmem38a | -0.032 |
| CABZ01074203.1 | 0.117 | celf2 | -0.032 |
| fgf10a | 0.110 | eno1a | -0.032 |
| FP325124.1 | 0.104 | gatm | -0.032 |
| fbln1 | 0.104 | ckmb | -0.032 |
| LOC101887111 | 0.104 | atp2a1 | -0.032 |
| CABZ01074130.1 | 0.101 | si:dkey-16p21.8 | -0.032 |
| col4a4 | 0.101 | klhl41b | -0.031 |
| LOC110438288 | 0.099 | ckma | -0.031 |
| CABZ01012962.1 | 0.098 | mylpfa | -0.031 |
| mknk2a | 0.098 | sparc | -0.031 |
| apoc1 | 0.095 | tpma | -0.031 |
| CABZ01067151.2 | 0.092 | tnnc2 | -0.030 |
| CU861477.1 | 0.090 | neb | -0.030 |
| vimr2 | 0.090 | acta1b | -0.030 |
| LOC101883748 | 0.089 | cox17 | -0.030 |
| LOC100538221 | 0.088 | acta1a | -0.029 |
| si:ch211-165f21.7 | 0.088 | ldb3a | -0.029 |
| hoxc12b | 0.088 | mybphb | -0.029 |
| nid2a | 0.087 | desma | -0.029 |
| her7 | 0.087 | vwde | -0.029 |
| itm2cb | 0.085 | gamt | -0.029 |
| dnase1l4.1 | 0.085 | pvalb1 | -0.029 |
| fgd5b | 0.084 | zgc:101853 | -0.029 |
| aipl1 | 0.083 | myl1 | -0.029 |
| XLOC-001600 | 0.081 | gapdh | -0.029 |
| col4a3 | 0.081 | CABZ01078594.1 | -0.029 |