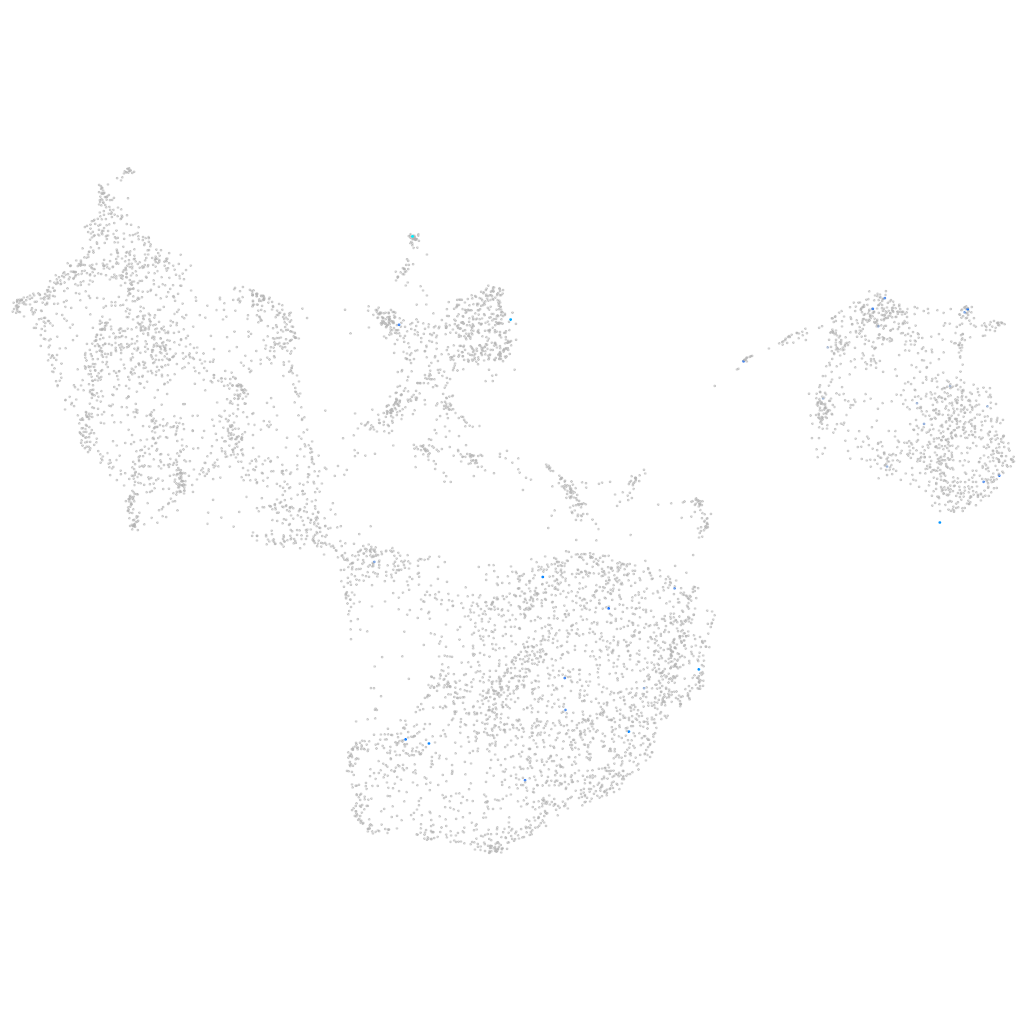
si:cabz01023700.1
ZFIN 
Other cell groups


all cells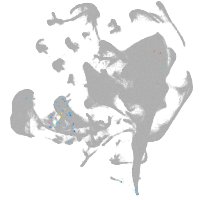 |
blastomeres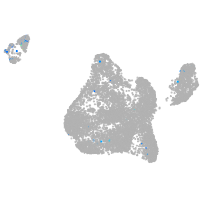 |
PGCs |
endoderm |
axial mesoderm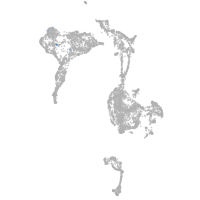 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory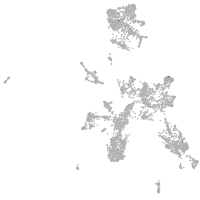 |
otic / lateral line |
epidermis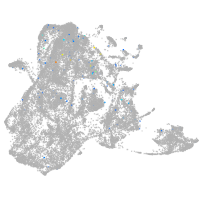 |
periderm |
fin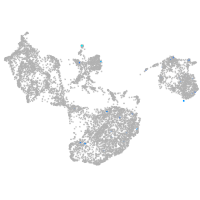 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC100331665 | 0.436 | c1qtnf5 | -0.034 |
| lgals9l4 | 0.436 | bhmt | -0.034 |
| si:dkey-14o18.2 | 0.416 | fbln1 | -0.032 |
| nrtn | 0.359 | hgd | -0.031 |
| zgc:193593 | 0.325 | khdrbs1a | -0.031 |
| cldn2 | 0.324 | cd82a | -0.030 |
| LOC100005908 | 0.309 | ecrg4a | -0.030 |
| LOC110439879 | 0.309 | twist1a | -0.029 |
| ttc8.1 | 0.285 | crabp2a | -0.029 |
| ptafr | 0.271 | pmp22a | -0.029 |
| hbaa2 | 0.267 | tmem119b | -0.029 |
| si:dkey-26i13.8 | 0.257 | pcbd1 | -0.029 |
| LOC103908672 | 0.245 | msna | -0.028 |
| XLOC-019884 | 0.239 | prrx1a | -0.028 |
| glis3 | 0.231 | prrx1b | -0.027 |
| BX927218.1 | 0.215 | dcn | -0.027 |
| LOC101882020 | 0.213 | fah | -0.027 |
| clul1 | 0.208 | ntd5 | -0.027 |
| rp1 | 0.200 | mfap2 | -0.027 |
| kcns3a | 0.199 | h2afva | -0.027 |
| rgs9a | 0.194 | gpc1a | -0.027 |
| lcp1 | 0.188 | cfl1 | -0.027 |
| lrit2 | 0.187 | ptx3a | -0.027 |
| XLOC-043546 | 0.186 | ttc36 | -0.027 |
| pde6h | 0.182 | qdpra | -0.027 |
| camk1gb | 0.182 | rpl22l1 | -0.027 |
| zgc:194686 | 0.178 | sept15 | -0.027 |
| cnga3b | 0.175 | reck | -0.026 |
| camk1ga | 0.166 | snrpe | -0.026 |
| BX119915.2 | 0.161 | cd81a | -0.026 |
| tdrd7b | 0.161 | ap1s2 | -0.025 |
| CU651662.1 | 0.154 | hoxa13b | -0.025 |
| guca1c | 0.152 | hpdb | -0.025 |
| inaa | 0.152 | hmgb3a | -0.025 |
| XLOC-035229 | 0.151 | pdgfra | -0.025 |