shroom family member 4
ZFIN 
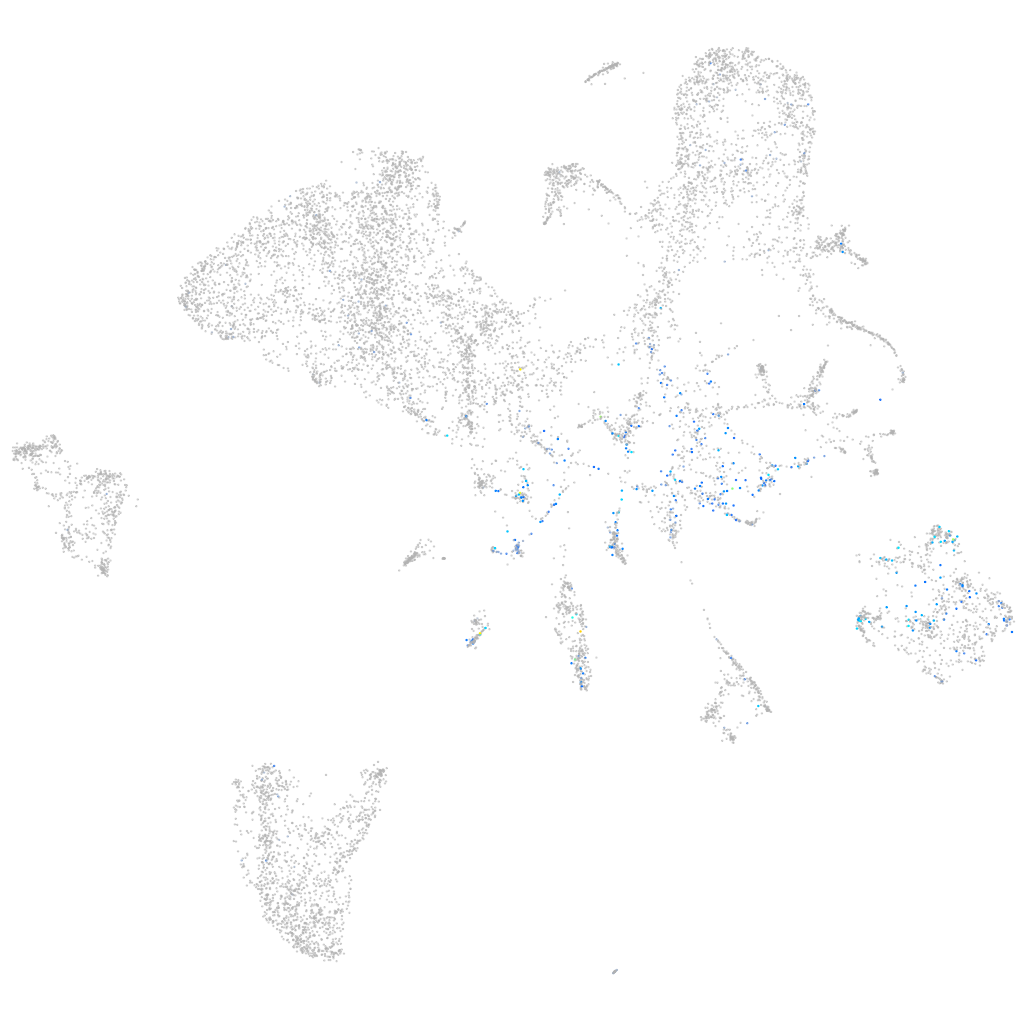
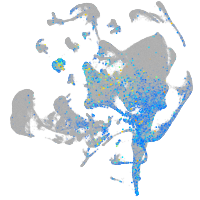
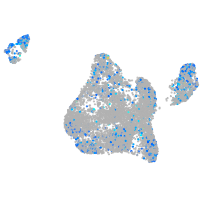
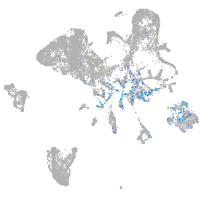
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tpm4a | 0.206 | gapdh | -0.148 |
| cx43.4 | 0.188 | ahcy | -0.144 |
| si:ch211-222l21.1 | 0.188 | eno3 | -0.143 |
| qkia | 0.186 | sod1 | -0.139 |
| hmgn6 | 0.185 | gamt | -0.136 |
| CABZ01075068.1 | 0.181 | nupr1b | -0.135 |
| apela | 0.179 | glud1b | -0.127 |
| marcksb | 0.178 | atp5if1b | -0.127 |
| cdh6 | 0.177 | aldob | -0.126 |
| khdrbs1a | 0.174 | gatm | -0.126 |
| oc90 | 0.174 | scp2a | -0.123 |
| h3f3d | 0.173 | dap | -0.122 |
| lsp1 | 0.172 | cx32.3 | -0.121 |
| hdac1 | 0.171 | suclg1 | -0.121 |
| hmgb3a | 0.170 | mat1a | -0.120 |
| nucks1a | 0.170 | tpi1b | -0.120 |
| si:ch73-281n10.2 | 0.170 | aldh7a1 | -0.118 |
| hnrnpaba | 0.169 | gstt1a | -0.117 |
| zbtb16a | 0.168 | fbp1b | -0.116 |
| fgfrl1b | 0.167 | pgk1 | -0.116 |
| syncrip | 0.167 | abat | -0.115 |
| hmga1a | 0.166 | gpx4a | -0.112 |
| smo | 0.166 | atp5l | -0.112 |
| marcksl1b | 0.165 | agxtb | -0.112 |
| akap12b | 0.165 | aldh6a1 | -0.111 |
| col8a1a | 0.164 | atp5mc1 | -0.111 |
| hspb1 | 0.163 | mdh1aa | -0.110 |
| znfl2a | 0.161 | apoa1b | -0.109 |
| cirbpb | 0.161 | gstp1 | -0.109 |
| cbx3a | 0.161 | eef1da | -0.108 |
| seta | 0.161 | atp5pf | -0.108 |
| h2afvb | 0.161 | sod2 | -0.107 |
| hmgn7 | 0.159 | ugt1a7 | -0.107 |
| sept15 | 0.159 | apoa4b.1 | -0.107 |
| id1 | 0.159 | gnmt | -0.107 |