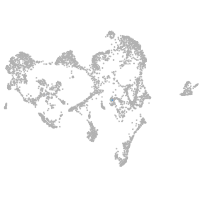
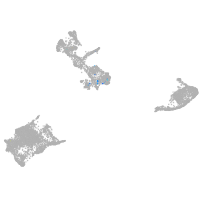
shisa family member 9b
ZFIN 
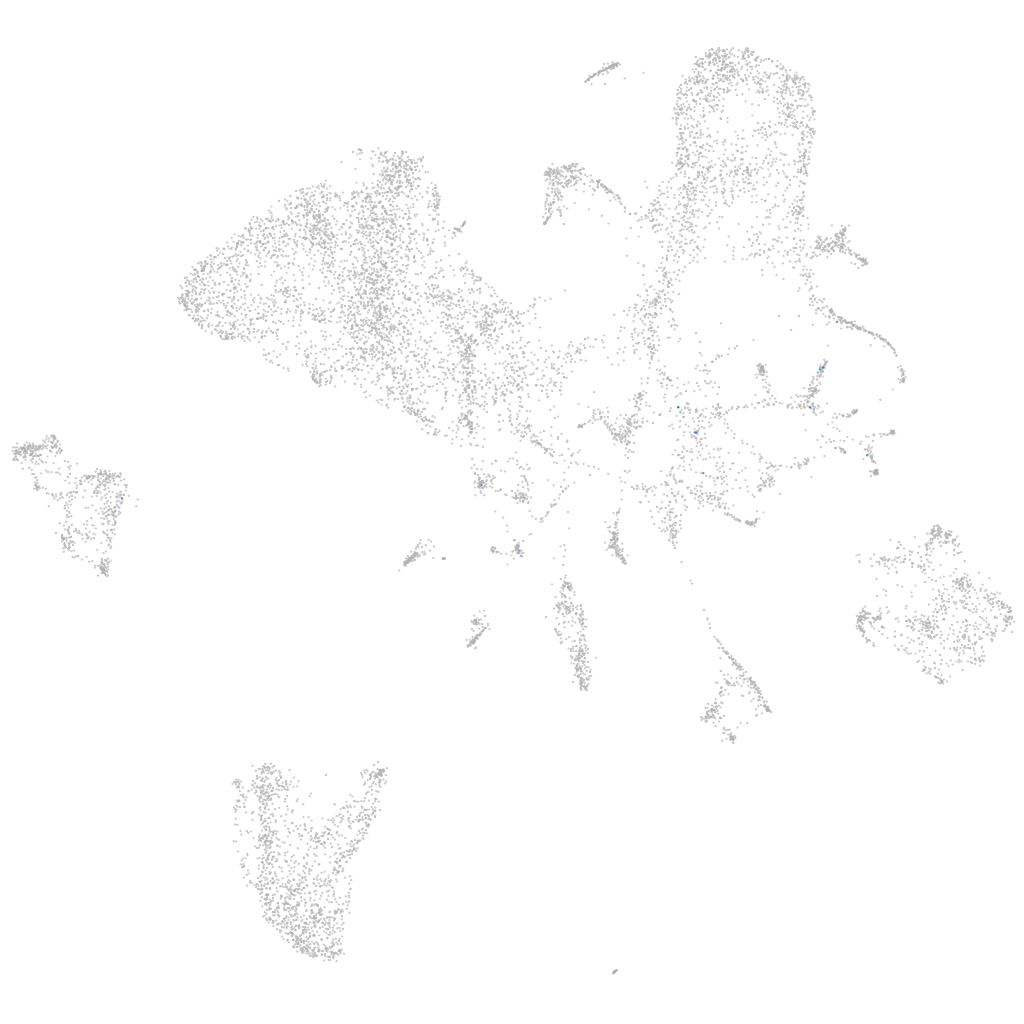
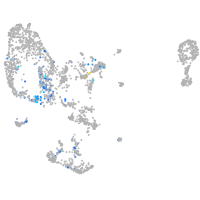
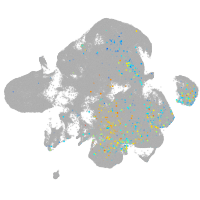
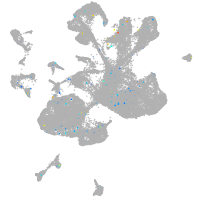
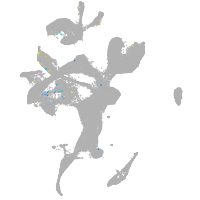
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcdh2g2 | 0.272 | gapdh | -0.043 |
| XLOC-007225 | 0.264 | ahcy | -0.040 |
| si:ch211-203b8.6 | 0.250 | gatm | -0.040 |
| si:dkey-260g12.1 | 0.245 | fbp1b | -0.035 |
| LOC110439381 | 0.241 | gamt | -0.034 |
| fndc4a | 0.237 | bhmt | -0.034 |
| BX936317.1 | 0.230 | apoc2 | -0.033 |
| LOC110439893 | 0.225 | apoa4b.1 | -0.033 |
| rxfp3 | 0.215 | mat1a | -0.031 |
| or137-4 | 0.215 | cx32.3 | -0.031 |
| si:ch211-154c21.1 | 0.215 | afp4 | -0.031 |
| CABZ01050238.1 | 0.214 | apoc1 | -0.030 |
| scpp1 | 0.212 | glud1b | -0.030 |
| gabra3 | 0.205 | cat | -0.030 |
| CT573467.1 | 0.205 | scp2a | -0.030 |
| LOC101885431 | 0.204 | apoa1b | -0.029 |
| rad21b | 0.190 | gstt1a | -0.029 |
| BX005368.1 | 0.177 | fabp2 | -0.029 |
| LOC110438150 | 0.175 | ugt1a7 | -0.029 |
| CT025909.1 | 0.171 | agxtb | -0.028 |
| CR457444.4 | 0.171 | cdo1 | -0.028 |
| CABZ01030278.1 | 0.171 | apobb.1 | -0.028 |
| LO018380.1 | 0.167 | chpt1 | -0.028 |
| trim55b | 0.165 | mt-nd4 | -0.028 |
| gpr22b | 0.162 | dhdhl | -0.028 |
| theg | 0.154 | dap | -0.028 |
| CU570976.1 | 0.153 | pla2g12b | -0.027 |
| chrna6 | 0.149 | msrb2 | -0.027 |
| htr2b | 0.144 | suclg2 | -0.027 |
| si:ch73-46n24.1 | 0.142 | slc27a2a | -0.027 |
| nrn1b | 0.141 | tdo2a | -0.027 |
| PRLHR | 0.138 | tfa | -0.027 |
| uts1 | 0.137 | eno3 | -0.027 |
| slitrk2 | 0.135 | mt-atp6 | -0.026 |
| gabrg1 | 0.135 | cth | -0.026 |