SH3-domain GRB2-like endophilin B2b
ZFIN 
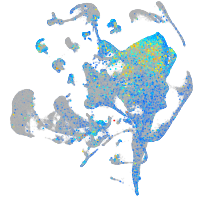
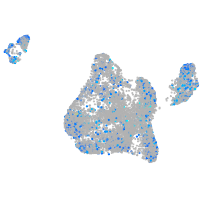
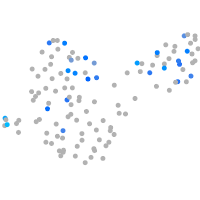
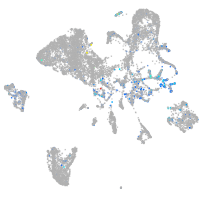
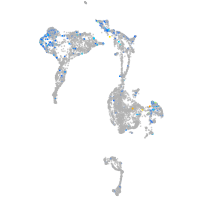
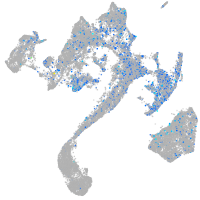
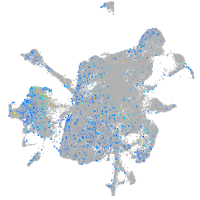
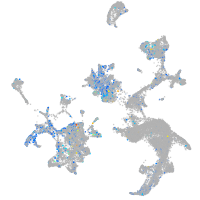
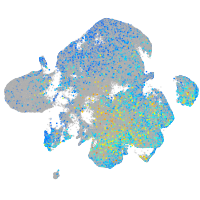
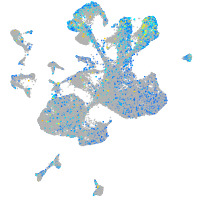
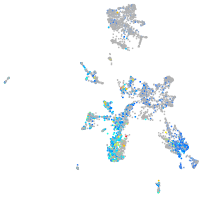
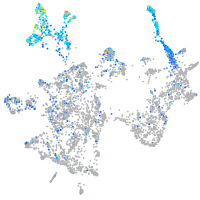
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.254 | gapdh | -0.158 |
| pax6b | 0.239 | gamt | -0.150 |
| neurod1 | 0.235 | ahcy | -0.145 |
| mir7a-1 | 0.228 | gatm | -0.143 |
| mir375-2 | 0.216 | fbp1b | -0.139 |
| gdi1 | 0.205 | bhmt | -0.125 |
| insm1a | 0.203 | nupr1b | -0.124 |
| rnasekb | 0.202 | mat1a | -0.124 |
| vat1 | 0.199 | glud1b | -0.124 |
| gnao1a | 0.198 | apoc2 | -0.123 |
| syt1a | 0.196 | apoa4b.1 | -0.122 |
| pcsk1nl | 0.194 | cx32.3 | -0.122 |
| vamp2 | 0.192 | aldh6a1 | -0.120 |
| lysmd2 | 0.192 | aldob | -0.118 |
| spire1b | 0.190 | apoa1b | -0.116 |
| elovl1a | 0.190 | pnp4b | -0.116 |
| dpysl2b | 0.190 | agxtb | -0.115 |
| pax4 | 0.188 | afp4 | -0.114 |
| scgn | 0.186 | eno3 | -0.113 |
| rprmb | 0.186 | mgst1.2 | -0.113 |
| kcnj11 | 0.185 | aqp12 | -0.111 |
| id4 | 0.185 | gstt1a | -0.111 |
| gpc1a | 0.184 | scp2a | -0.110 |
| cnrip1a | 0.183 | gcshb | -0.109 |
| myt1b | 0.183 | apobb.1 | -0.109 |
| tmsb | 0.183 | aldh7a1 | -0.108 |
| fev | 0.182 | cdo1 | -0.107 |
| gapdhs | 0.179 | hao1 | -0.107 |
| cplx2l | 0.178 | aldh1l1 | -0.106 |
| tox | 0.178 | apoa2 | -0.105 |
| si:dkey-153k10.9 | 0.177 | agxta | -0.105 |
| jagn1a | 0.177 | serpina1l | -0.104 |
| LOC110438555 | 0.176 | slc27a2a | -0.104 |
| syt11a | 0.173 | gpx4a | -0.104 |
| atp6v0cb | 0.172 | abat | -0.103 |