serum/glucocorticoid regulated kinase family member 3
ZFIN 
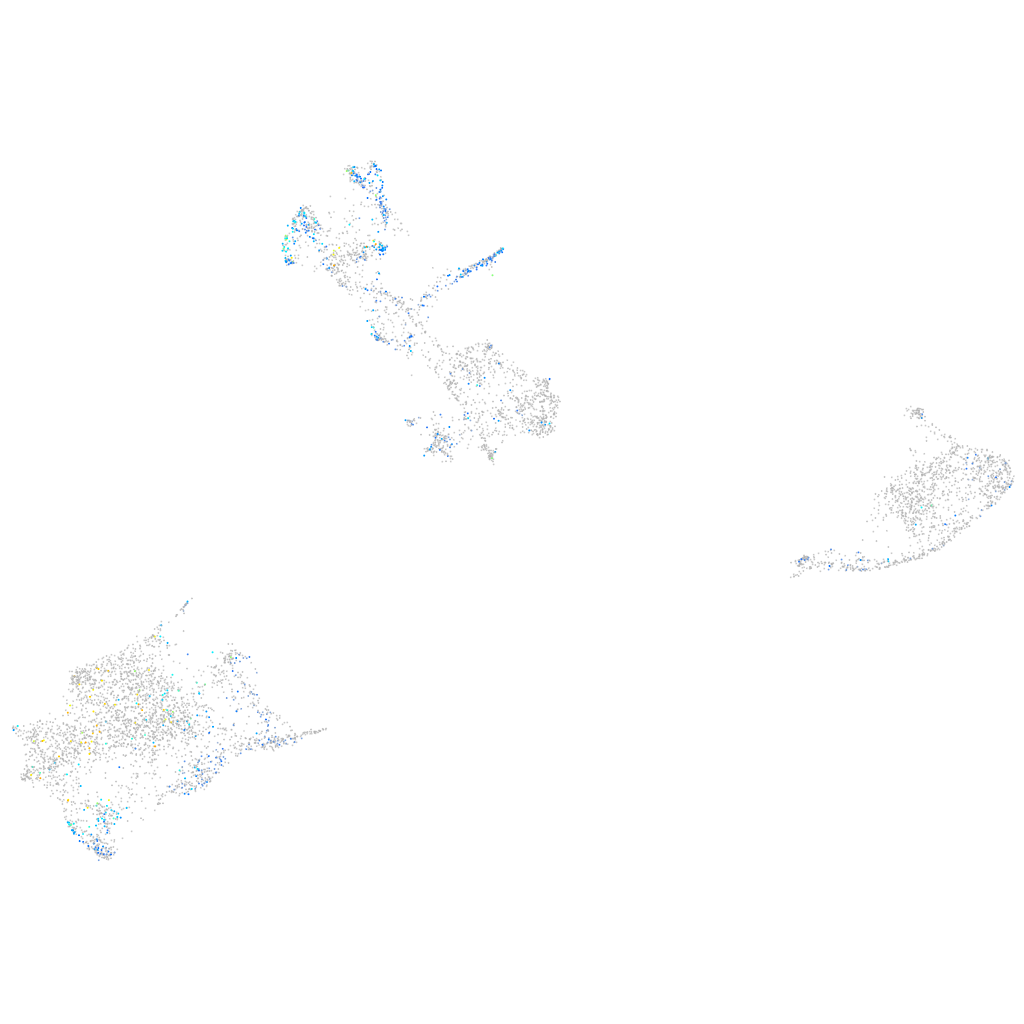
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-256m1.8 | 0.215 | pmela | -0.116 |
| gpnmb | 0.207 | tyrp1b | -0.113 |
| unm-sa821 | 0.202 | zgc:91968 | -0.110 |
| apoda.1 | 0.201 | tyrp1a | -0.103 |
| lypc | 0.198 | dct | -0.103 |
| defbl1 | 0.196 | oca2 | -0.092 |
| rgs4 | 0.194 | gstt1a | -0.092 |
| alx4b | 0.193 | qdpra | -0.091 |
| si:ch73-89b15.3 | 0.190 | tyr | -0.088 |
| prx | 0.190 | prkar1b | -0.081 |
| sytl2b | 0.190 | tuba1a | -0.080 |
| guk1a | 0.188 | si:ch211-222l21.1 | -0.079 |
| akap12a | 0.188 | anxa1a | -0.078 |
| sort1a | 0.182 | slc24a5 | -0.078 |
| pnp4a | 0.180 | gch2 | -0.075 |
| ltk | 0.180 | pcbd1 | -0.073 |
| tcirg1a | 0.179 | nova2 | -0.072 |
| s100v2 | 0.179 | mtbl | -0.072 |
| CABZ01072077.1 | 0.179 | kcnj13 | -0.072 |
| tmem179b | 0.177 | kita | -0.072 |
| sytl2a | 0.177 | cotl1 | -0.068 |
| alx4a | 0.176 | tuba1c | -0.068 |
| zgc:110789 | 0.176 | lamp1a | -0.067 |
| cmtm6 | 0.175 | si:ch73-389b16.1 | -0.066 |
| csf1a | 0.175 | tmsb | -0.066 |
| si:dkey-197i20.6 | 0.175 | pah | -0.065 |
| slc25a36a | 0.174 | aadac | -0.063 |
| alx1 | 0.173 | stmn1b | -0.063 |
| clec3ba | 0.170 | marcksb | -0.062 |
| slc22a31 | 0.167 | elavl3 | -0.061 |
| tmem63bb | 0.166 | slc22a2 | -0.059 |
| syne1b | 0.163 | gpm6aa | -0.058 |
| tfec | 0.162 | yjefn3 | -0.058 |
| pltp | 0.162 | tfap2e | -0.057 |
| si:ch211-105c13.3 | 0.161 | timeless | -0.057 |