"sarcoglycan, epsilon"
ZFIN 
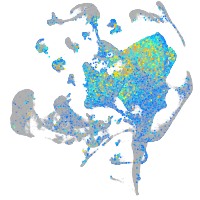
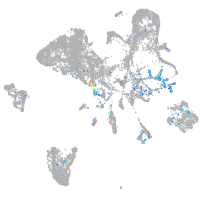
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.265 | gapdh | -0.128 |
| syt1a | 0.259 | gamt | -0.124 |
| vat1 | 0.237 | ahcy | -0.123 |
| mir7a-1 | 0.236 | aldob | -0.123 |
| neurod1 | 0.234 | gatm | -0.120 |
| rnasekb | 0.233 | hdlbpa | -0.118 |
| vamp2 | 0.224 | fbp1b | -0.116 |
| pcsk1nl | 0.222 | gstt1a | -0.114 |
| lysmd2 | 0.216 | aldh6a1 | -0.110 |
| si:dkey-153k10.9 | 0.212 | cx32.3 | -0.104 |
| tspan7b | 0.212 | nupr1b | -0.102 |
| cplx2l | 0.209 | mat1a | -0.100 |
| jpt1b | 0.208 | dap | -0.098 |
| gnao1a | 0.207 | eno3 | -0.097 |
| mllt11 | 0.204 | nipsnap3a | -0.096 |
| slc35g2b | 0.204 | apoa4b.1 | -0.096 |
| atp6v0cb | 0.202 | aldh7a1 | -0.095 |
| fez1 | 0.202 | scp2a | -0.095 |
| scgn | 0.199 | pck2 | -0.095 |
| gng3 | 0.199 | suclg1 | -0.094 |
| aldocb | 0.199 | aqp12 | -0.094 |
| egr4 | 0.198 | abat | -0.094 |
| atpv0e2 | 0.197 | sult2st2 | -0.093 |
| kcnj11 | 0.197 | haao | -0.093 |
| syt11a | 0.196 | gstr | -0.093 |
| id4 | 0.196 | ugt1a7 | -0.093 |
| tox | 0.195 | rpl4 | -0.093 |
| gapdhs | 0.195 | agxtb | -0.092 |
| stmn1b | 0.194 | slc25a34 | -0.092 |
| insm1a | 0.194 | gnmt | -0.092 |
| jagn1a | 0.193 | suclg2 | -0.092 |
| LOC100537384 | 0.193 | tdo2a | -0.091 |
| cnrip1a | 0.192 | pnp4b | -0.091 |
| tuba1c | 0.192 | glud1b | -0.091 |
| myt1b | 0.192 | mgst1.2 | -0.091 |