"sarcoglycan, alpha"
ZFIN 
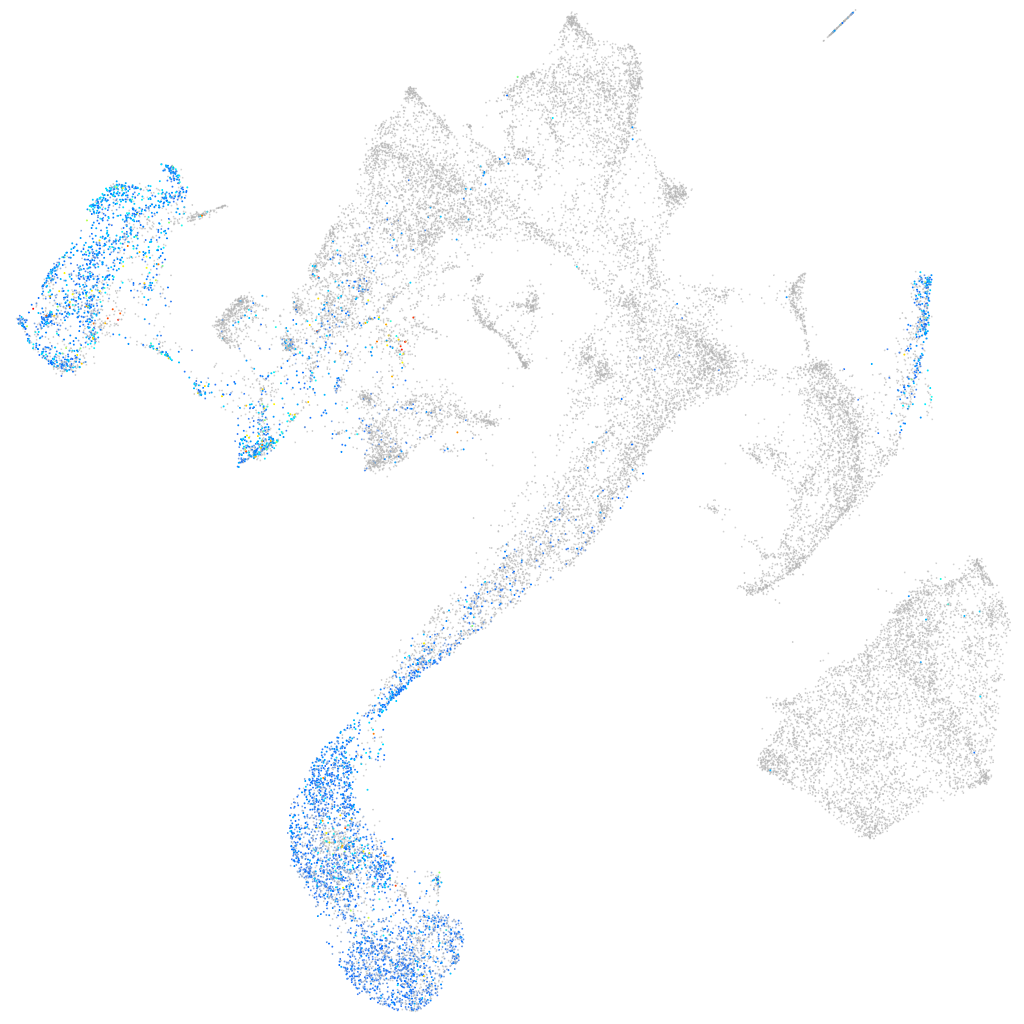
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| casq2 | 0.577 | hsp90ab1 | -0.524 |
| neb | 0.574 | pabpc1a | -0.474 |
| actn3b | 0.557 | h3f3d | -0.470 |
| aldoab | 0.553 | si:ch73-1a9.3 | -0.464 |
| atp2a1 | 0.552 | ran | -0.462 |
| cav3 | 0.551 | hmgb2b | -0.457 |
| srl | 0.551 | khdrbs1a | -0.452 |
| si:ch211-266g18.10 | 0.550 | h2afvb | -0.449 |
| tmem38a | 0.545 | cirbpb | -0.449 |
| ak1 | 0.545 | hnrnpaba | -0.438 |
| cavin4b | 0.544 | si:ch211-222l21.1 | -0.434 |
| ldb3b | 0.543 | ptmab | -0.424 |
| ckmb | 0.542 | hmgn2 | -0.423 |
| ckma | 0.541 | hnrnpabb | -0.418 |
| gapdh | 0.540 | hmgb2a | -0.414 |
| ttn.2 | 0.539 | cirbpa | -0.410 |
| trdn | 0.537 | hnrnpa0b | -0.406 |
| cavin4a | 0.536 | setb | -0.405 |
| myom1a | 0.533 | hmga1a | -0.404 |
| desma | 0.533 | cbx3a | -0.403 |
| actc1b | 0.533 | ubc | -0.403 |
| pgam2 | 0.529 | hmgn7 | -0.399 |
| actn3a | 0.528 | si:ch73-281n10.2 | -0.398 |
| ldb3a | 0.528 | tuba8l4 | -0.389 |
| smyd1a | 0.527 | naca | -0.389 |
| xirp2a | 0.527 | fthl27 | -0.387 |
| eno3 | 0.524 | actb2 | -0.386 |
| tnnc2 | 0.522 | snrpf | -0.382 |
| tmem182a | 0.521 | cfl1 | -0.379 |
| CABZ01078594.1 | 0.519 | anp32b | -0.377 |
| tmod4 | 0.517 | sumo3a | -0.376 |
| ttn.1 | 0.516 | snrpd1 | -0.376 |
| prx | 0.515 | si:ch211-288g17.3 | -0.374 |
| klhl43 | 0.512 | eef1a1l1 | -0.373 |
| myom1b | 0.508 | syncrip | -0.373 |