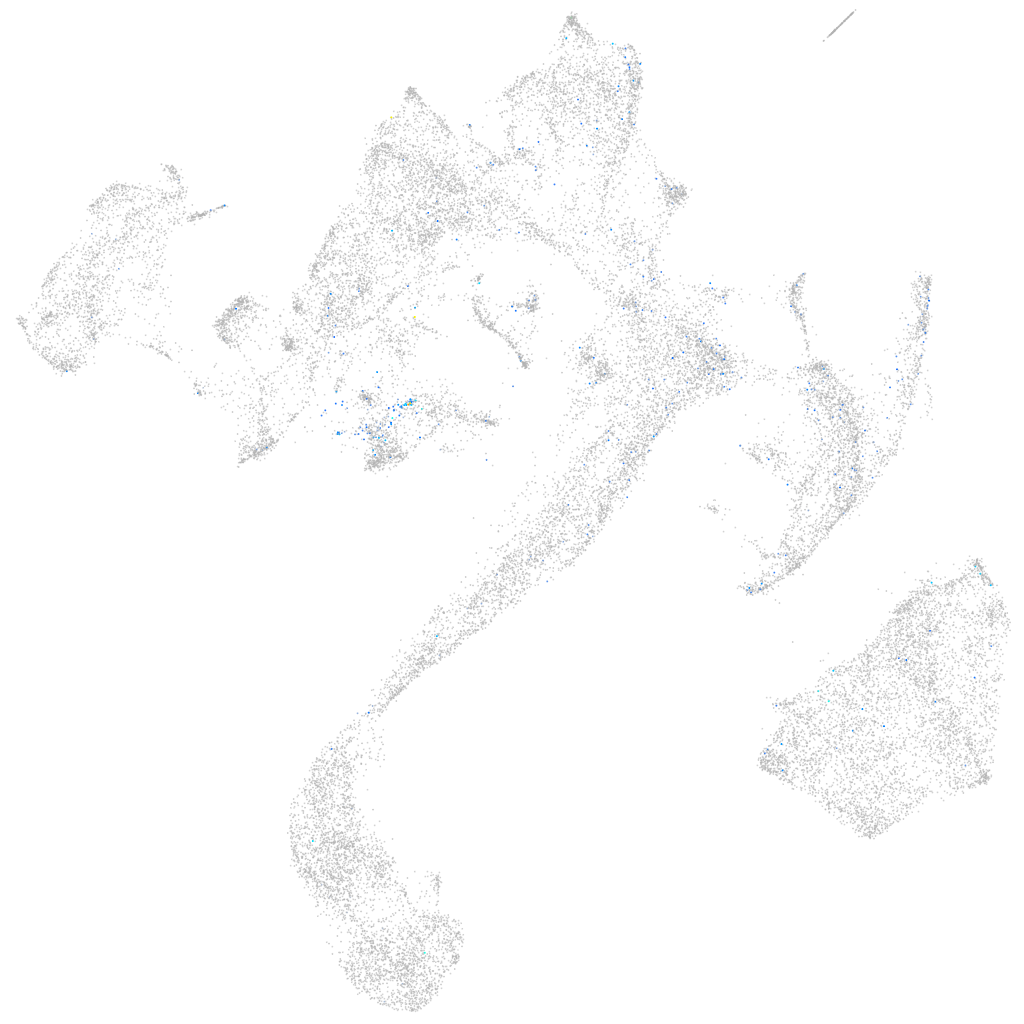
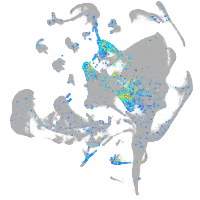
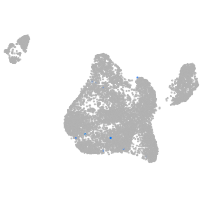
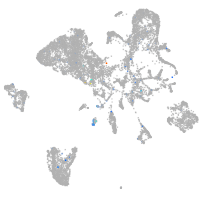
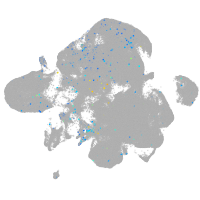
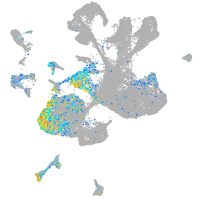
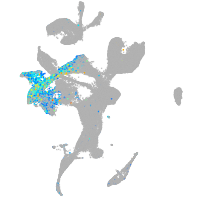
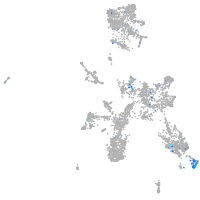
septin 8b
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr37l1b | 0.306 | si:ch73-367p23.2 | -0.036 |
| si:ch211-66e2.5 | 0.301 | ank1a | -0.035 |
| slc6a11b | 0.281 | ckma | -0.035 |
| slc1a2b | 0.272 | XLOC-025819 | -0.035 |
| atp1a1b | 0.261 | ldb3b | -0.035 |
| si:ch211-180a12.2 | 0.259 | XLOC-001975 | -0.035 |
| FO704813.1 | 0.235 | XLOC-006515 | -0.034 |
| hepacama | 0.220 | pgam2 | -0.034 |
| slc1a3b | 0.200 | acta1b | -0.034 |
| cx43 | 0.188 | myom1a | -0.034 |
| slc6a1b | 0.183 | XLOC-005350 | -0.034 |
| fabp7a | 0.182 | tnnc2 | -0.034 |
| eno1b | 0.177 | CR855257.1 | -0.034 |
| zgc:165461 | 0.173 | atp2a1 | -0.033 |
| slc7a10a | 0.171 | aldoab | -0.033 |
| slc4a4a | 0.170 | actn3a | -0.033 |
| s100b | 0.167 | gapdh | -0.033 |
| ptn | 0.166 | tmod4 | -0.033 |
| si:dkey-245n4.2 | 0.164 | mybphb | -0.033 |
| slc38a3a | 0.161 | jph2 | -0.033 |
| si:ch211-106h11.1 | 0.158 | CABZ01061524.1 | -0.033 |
| cd81b | 0.155 | ryr1b | -0.032 |
| wnt7aa | 0.155 | neb | -0.032 |
| meltf | 0.145 | myoz1b | -0.032 |
| cspg5b | 0.140 | eef2l2 | -0.032 |
| kcnj10a | 0.140 | ckmb | -0.032 |
| CU467822.1 | 0.133 | dhrs7cb | -0.032 |
| atp1b4 | 0.131 | casq1b | -0.032 |
| acana | 0.130 | tnnt3a | -0.032 |
| matn1 | 0.129 | cfl2 | -0.032 |
| ca4a | 0.127 | smyd1a | -0.031 |
| mia | 0.126 | XLOC-030027 | -0.031 |
| crtac1a | 0.126 | hacd1 | -0.031 |
| hspb15 | 0.124 | tmem38a | -0.031 |
| prss35 | 0.123 | CABZ01078594.1 | -0.031 |