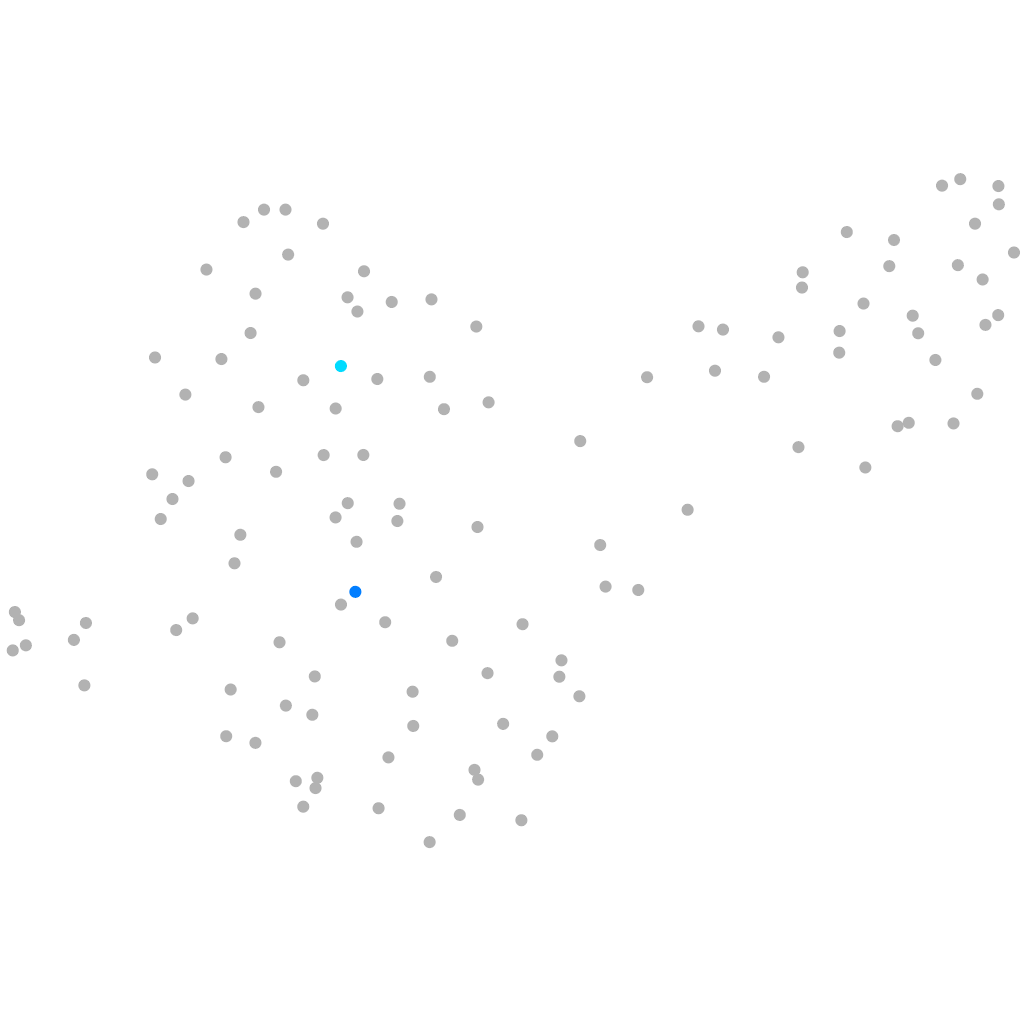
septin 4b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm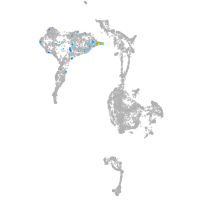 |
muscle 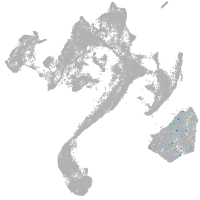 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 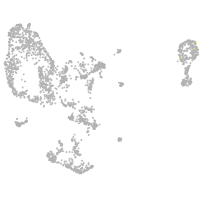 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| efr3ba | 0.864 | nme2b.1 | -0.233 |
| emilin2a | 0.864 | rpl39 | -0.224 |
| kif3cb | 0.864 | rps25 | -0.219 |
| si:ch1073-513e17.1 | 0.864 | rpl13 | -0.212 |
| si:dkey-1h24.2 | 0.864 | slc25a5 | -0.178 |
| cenpv | 0.788 | cox4i1 | -0.172 |
| ugt1b5 | 0.778 | rps24 | -0.172 |
| LOC108181079 | 0.743 | sub1b | -0.163 |
| CU695223.1 | 0.737 | strap | -0.163 |
| pnp5b | 0.736 | tegt | -0.162 |
| hoxa13b | 0.735 | ipo7 | -0.160 |
| XLOC-025146 | 0.733 | atp5pb | -0.160 |
| LOC108181166 | 0.728 | rpl29 | -0.157 |
| kita | 0.708 | pdcd4b | -0.156 |
| CT025771.1 | 0.706 | ssrp1a | -0.152 |
| exoc3l1 | 0.699 | paics | -0.151 |
| RNF122 | 0.683 | rps21 | -0.150 |
| fam171b | 0.674 | rpl36a | -0.149 |
| pdss2 | 0.661 | copz1 | -0.149 |
| pdlim4 | 0.657 | vdac3 | -0.148 |
| arhgap36 | 0.655 | eno3 | -0.147 |
| filip1l | 0.653 | sap18 | -0.147 |
| si:ch73-89b15.3 | 0.646 | eif3m | -0.145 |
| LOC100537032 | 0.643 | snrpa1 | -0.145 |
| LOC100535210 | 0.638 | arf2b | -0.144 |
| disp1 | 0.629 | ywhaz | -0.143 |
| trim62 | 0.625 | slc7a6os | -0.143 |
| exoc3 | 0.623 | tmsb4x | -0.143 |
| bin1a | 0.616 | myl12.1 | -0.142 |
| ano1 | 0.599 | arpc3 | -0.139 |
| LOC101884382 | 0.591 | cst14a.2 | -0.139 |
| LOC110438587 | 0.586 | psmb4 | -0.139 |
| slc4a1b | 0.580 | ythdf2 | -0.138 |
| camkva | 0.574 | cd9b | -0.135 |
| znf692 | 0.571 | emc10 | -0.135 |