"sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Aa"
ZFIN 
Other cell groups


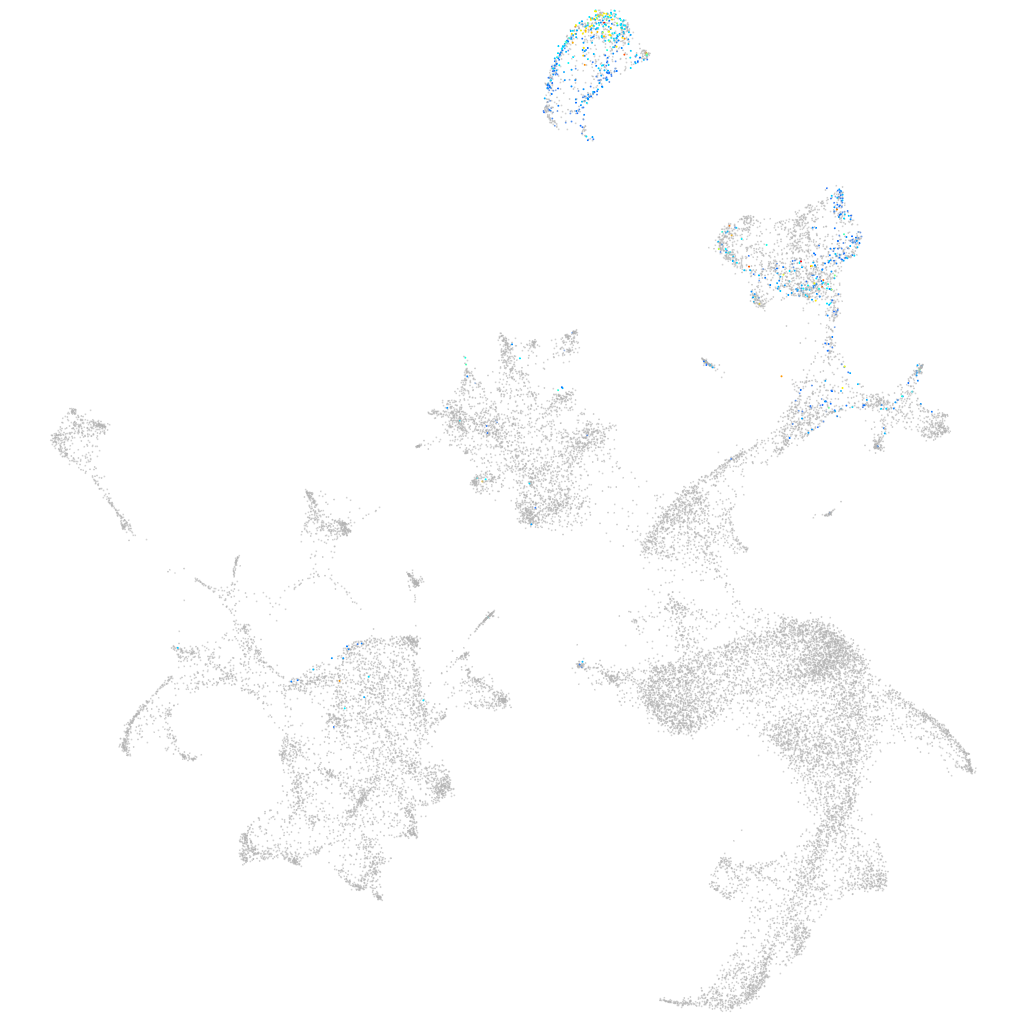
all cells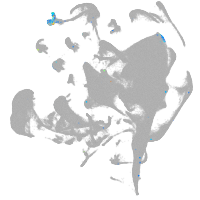 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 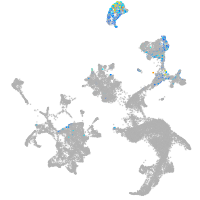 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cfbl | 0.430 | hbae1.1 | -0.157 |
| lect2l | 0.426 | hbae3 | -0.153 |
| si:dkey-102g19.3 | 0.425 | hbbe1.3 | -0.152 |
| mmp13a | 0.421 | hbae1.3 | -0.151 |
| glipr1a | 0.419 | hbbe2 | -0.149 |
| alox5ap | 0.398 | hbbe1.1 | -0.143 |
| npsn | 0.397 | cahz | -0.135 |
| ponzr6 | 0.394 | hbbe1.2 | -0.130 |
| mpx | 0.393 | hemgn | -0.128 |
| scinlb | 0.390 | blvrb | -0.123 |
| lta4h | 0.384 | zgc:56095 | -0.122 |
| il1b | 0.384 | fth1a | -0.121 |
| fcer1gl | 0.380 | alas2 | -0.120 |
| spice1 | 0.377 | si:ch211-250g4.3 | -0.119 |
| il34 | 0.375 | lmo2 | -0.117 |
| scpp8 | 0.374 | creg1 | -0.114 |
| ctss1 | 0.374 | slc4a1a | -0.112 |
| lyz | 0.369 | nt5c2l1 | -0.110 |
| ch25hl2 | 0.368 | zgc:163057 | -0.106 |
| ctss2.1 | 0.365 | epb41b | -0.106 |
| spi1b | 0.360 | si:ch211-207c6.2 | -0.102 |
| si:ch211-276a23.5 | 0.358 | nmt1b | -0.097 |
| itgb2 | 0.356 | tmod4 | -0.093 |
| si:ch211-193e13.5 | 0.355 | aqp1a.1 | -0.092 |
| LOC101884267 | 0.355 | plac8l1 | -0.092 |
| si:ch1073-443f11.2 | 0.355 | prdx2 | -0.091 |
| cpa5 | 0.355 | sptb | -0.086 |
| CABZ01001434.1 | 0.351 | hbae5 | -0.086 |
| coro1a | 0.348 | si:ch211-227m13.1 | -0.085 |
| BX908782.3 | 0.347 | uros | -0.085 |
| si:ch73-343l4.8 | 0.346 | znfl2a | -0.083 |
| dusp2 | 0.344 | tfr1a | -0.083 |
| dhrs9 | 0.340 | tspo | -0.081 |
| si:dkey-27h10.2 | 0.340 | hdr | -0.081 |
| gapdh | 0.340 | rfesd | -0.081 |