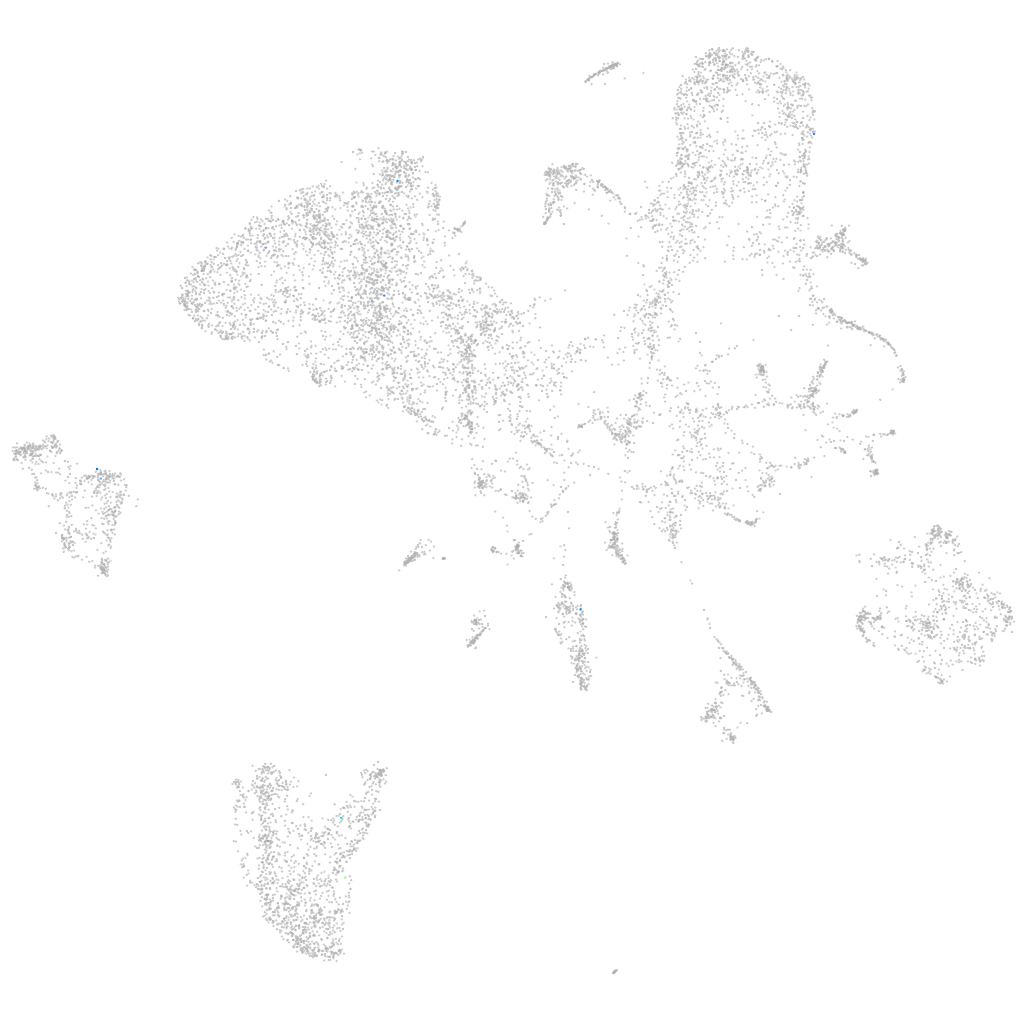
selenoprotein U1b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 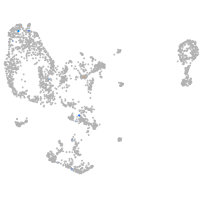 |
neural |
spinal cord / glia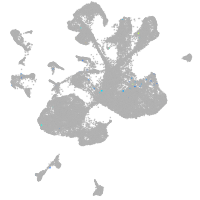 |
eye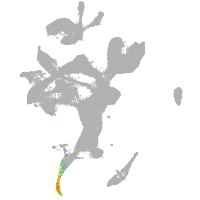 |
taste / olfactory |
otic / lateral line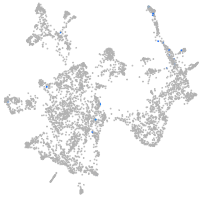 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU651662.1 | 0.504 | rps26 | -0.038 |
| BX005419.1 | 0.488 | faua | -0.030 |
| AL954142.2 | 0.477 | atp5mc3b | -0.029 |
| galnt18a | 0.379 | rps26l | -0.028 |
| pappa2 | 0.366 | uba52 | -0.027 |
| pde11al | 0.325 | rpl38 | -0.026 |
| inhbab | 0.310 | naca | -0.025 |
| pmelb | 0.307 | cox7c | -0.024 |
| si:ch211-106h4.12 | 0.303 | eif2s2 | -0.024 |
| tecta | 0.294 | rpl10a | -0.024 |
| si:dkey-57k2.6 | 0.268 | atp5md | -0.024 |
| mmel1 | 0.235 | atp5pb | -0.023 |
| olfml2a | 0.224 | tma7 | -0.023 |
| tyr | 0.219 | cox7b | -0.023 |
| jam2b | 0.207 | rps29 | -0.022 |
| acot21 | 0.206 | eif5 | -0.022 |
| shisa3 | 0.204 | khdrbs1a | -0.022 |
| slco1e1 | 0.202 | atp5meb | -0.022 |
| LOC100537649 | 0.200 | fth1a | -0.021 |
| dct | 0.198 | tuba8l4 | -0.021 |
| grm6b | 0.184 | rps3 | -0.021 |
| tyrp1b | 0.181 | snrpe | -0.021 |
| BX927333.1 | 0.180 | atp5mc3a | -0.020 |
| adamts15a | 0.179 | rps23 | -0.020 |
| ADAM12 (1 of many) | 0.176 | rpl22l1 | -0.020 |
| msnb | 0.176 | cox7a2a | -0.020 |
| sytl2a | 0.175 | erh | -0.020 |
| cidea | 0.174 | ywhabl | -0.019 |
| tspan10 | 0.171 | si:dkey-67c22.2 | -0.019 |
| satb1a | 0.166 | ube2ib | -0.019 |
| pald1a | 0.163 | rbm8a | -0.019 |
| loxl1 | 0.162 | pfn2l | -0.019 |
| LOC108182795 | 0.159 | si:dkey-151g10.6 | -0.019 |
| pcolcea | 0.159 | ndufa12 | -0.019 |
| si:ch211-106j24.1 | 0.157 | eif3f | -0.019 |