selectin E
ZFIN 
Other cell groups


all cells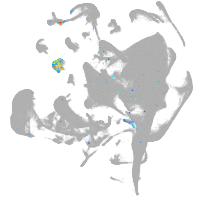 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 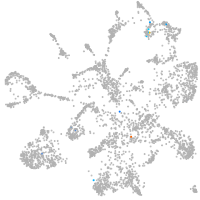 |
mesenchyme 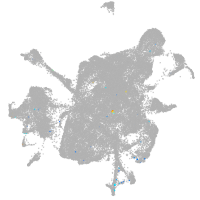 |
hematopoietic / vasculature 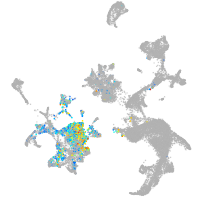 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr182 | 0.507 | hbae1.1 | -0.252 |
| plvapb | 0.484 | hbae1.3 | -0.244 |
| cpn1 | 0.454 | prdx2 | -0.244 |
| mrc1a | 0.453 | hbbe2 | -0.244 |
| si:ch211-145b13.6 | 0.449 | hbae3 | -0.238 |
| stab2 | 0.447 | hbbe1.3 | -0.234 |
| dab2 | 0.444 | hbbe1.1 | -0.230 |
| lyve1b | 0.441 | cahz | -0.226 |
| si:dkey-28n18.9 | 0.431 | hemgn | -0.214 |
| krt18a.1 | 0.421 | hbbe1.2 | -0.210 |
| cd81a | 0.417 | blvrb | -0.206 |
| XLOC-016094 | 0.415 | fth1a | -0.200 |
| krt8 | 0.412 | alas2 | -0.198 |
| cdh5 | 0.402 | tspo | -0.192 |
| aplnra | 0.400 | si:ch211-250g4.3 | -0.188 |
| tie1 | 0.397 | slc4a1a | -0.187 |
| prcp | 0.395 | gpx1a | -0.184 |
| stab1 | 0.394 | nt5c2l1 | -0.183 |
| ramp2 | 0.392 | creg1 | -0.176 |
| c1qtnf6a | 0.390 | zgc:163057 | -0.176 |
| il13ra2 | 0.389 | epb41b | -0.176 |
| lfng | 0.388 | si:ch211-207c6.2 | -0.169 |
| clec14a | 0.387 | nmt1b | -0.160 |
| fam174b | 0.382 | tmod4 | -0.152 |
| ccdc80 | 0.374 | plac8l1 | -0.150 |
| pecam1 | 0.373 | si:ch211-227m13.1 | -0.141 |
| sypl2a | 0.372 | hbae5 | -0.139 |
| fam198b | 0.371 | vdac2 | -0.139 |
| plk2b | 0.371 | tfr1a | -0.138 |
| si:dkey-261h17.1 | 0.370 | sptb | -0.135 |
| cyp1a | 0.361 | tdh | -0.134 |
| calcrlb | 0.359 | rfesd | -0.134 |
| rab11bb | 0.359 | tmem14ca | -0.133 |
| etv2 | 0.354 | znfl2a | -0.129 |
| tpm4a | 0.354 | uros | -0.127 |




