"SCY1-like, kinase-like 1"
ZFIN 
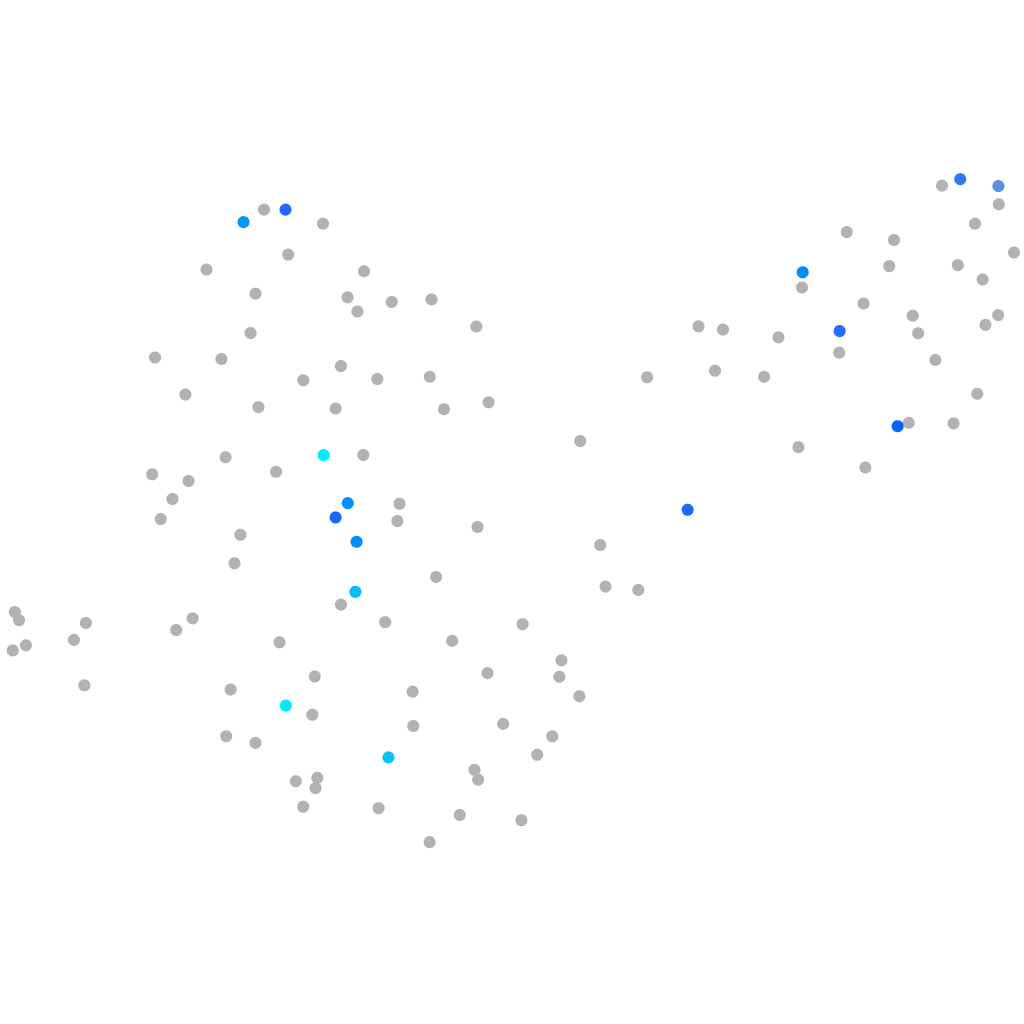
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sp8b | 0.510 | dynlrb1 | -0.218 |
| pi4kb | 0.479 | chmp5b | -0.210 |
| tcf19l | 0.476 | prdx2 | -0.209 |
| her8a | 0.437 | gng5 | -0.209 |
| zeb2b | 0.436 | si:dkey-276j7.1 | -0.206 |
| tpbga | 0.429 | rps14 | -0.198 |
| sgk494b | 0.425 | btg1 | -0.197 |
| arhgef39 | 0.425 | atp10a | -0.195 |
| slc38a3b | 0.424 | cst14a.2 | -0.192 |
| wipf1b | 0.423 | tomm20a | -0.190 |
| yme1l1a | 0.420 | tktb | -0.188 |
| sall1a | 0.417 | timm10b | -0.187 |
| cers5 | 0.414 | rnasekb | -0.186 |
| snx24 | 0.411 | eif4ba | -0.185 |
| TCIM | 0.411 | sptssa | -0.185 |
| LOC101882575 | 0.411 | si:dkeyp-104h9.5 | -0.184 |
| ptprt | 0.411 | polr2b | -0.183 |
| chadla | 0.401 | capzb | -0.183 |
| en1b | 0.401 | xpo7 | -0.182 |
| LOC101882072 | 0.401 | nme2b.1 | -0.181 |
| LOC101885180 | 0.401 | purba | -0.181 |
| mical2a | 0.401 | gstk1 | -0.179 |
| GNG4 | 0.400 | etfa | -0.179 |
| pim2 | 0.398 | nsa2 | -0.179 |
| tyro3 | 0.388 | ppid | -0.177 |
| mib1 | 0.388 | eno3 | -0.177 |
| slitrk3a | 0.387 | anapc15 | -0.177 |
| dlc | 0.386 | prdx4 | -0.176 |
| rc3h1b | 0.378 | copb2 | -0.176 |
| slc35a3b | 0.377 | fkbp7 | -0.175 |
| itgb1b | 0.375 | jpt1b | -0.175 |
| CABZ01072950.1 | 0.374 | clip1a | -0.174 |
| irgf4 | 0.371 | eif1ad | -0.174 |
| lrrc29 | 0.367 | cox5ab | -0.174 |
| admp | 0.365 | cox6c | -0.174 |